World Journal of AIDS
Vol. 3 No. 2 (2013) , Article ID: 32760 , 16 pages DOI:10.4236/wja.2013.32018
RNA Wave for the HIV Therapy: Foods, Stem Cells and the RNA Information Gene
![]()
Retroviral Genetics Group, Nagoya City University, Nagoya, Japan.
Email: fatfuji@hotmail.co.jp
Copyright © 2013 Yoichi Robertus Fujii. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received March 7th, 2013; revised April 6th, 2013; accepted April 14th, 2013
Keywords: CCR5∆32; HIV-1; Edible Vaccine; iPS cell; Microrna; RNA Information; RNA Wave, siRNA; Stem Cell Therapy
ABSTRACT
The microRNA (miRNA) gene is small RNA molecule, approximate 20 nucleotides (nts) in length, and also the miRNA is information in a cell as well as the mobile genetic information; therefore, when only one kind of tumor suppressor RNA information gene (Rig) was intravenously administrated, tumorigenic cells can be retuned to the normal cells in vivo. Although the processes of oncogenic have multiple ways, Rig can control its complex system, such as cell cycle with tuning to translation and transcription processing systems. In quite recent experiments, human breast milk and bovine milk have contained Rigs into their microvesicular components. Both also contain the infant nutrient elements. Further, the siRNA genes in artificial nanoparticles were delivered via oral and could restore mouse intestinal inflamemation. In general, Rigs in the diet were found stable to orally affect the digested animals, therefore, the xenotropic Rigs in Rig transgenic plants could also protect from HIV-1 infection by the edible vaccine via intestinal cells. Because orally delivered miRNA as information could be incorporated into intestinal cells and transmitted into intraand inter-cells and between individuals to wave the system of translation and transcription. Given these mobile characters of Rigs, even though there is the xenotropic miRNA issue, edible Rig agents in plants as a vaccine would be applicable for the Rig diseases (RigDs) by the information technology-based therapy (iTBT) cooperated with system-based therapies such as stem cell therapy and chemotherapy.
1. Introduction
For eradication of human immunodeficiency virus type 1 (HIV-1), one of transposable elements (TEs), we should recognize that the TEs might be one of the RNA genes and the TEs is major source of the microRNA (miRNA) gene in the genome [1]. The control of epigene is involved in the eradication of HIV-1 because provirus of HIV-1 is held in the human genome of the infected reservoir cells. Therefore, miRNAs and the TEs are implicated in gene and genome evolution [2-6]. Long-terminal repeat (LTR) retrotransposons are the most abundant TEs in the Plant kingdom [7]. In some plants, LTR retrotransposon can make up more than 70% of the genome [8], whereas in human genome the LTR transposons are more than 8% [1,9]. Although the retrotransposition in the human genome induces excessive gene expression near the insertion site and occasionally occurs tumorigenesis, it has recently been proved that the retrotransposon Alu insertion inactivates the neighboring progesterone receptor gene expression through an epigenetic mechanism [10]. DNA methylation is thought to be the mechanism; however, Byun et al. described that what triggers the DNA methylation is not clear, therefore, environmental cues are believed to be responsible for promoting movement of its transposon. This finding strongly suggested a possibility of the RNA wave that TE-derived many RNA information genes (Rigs), such as miRNA genes can trigger the change of the epigenetic state [1]. Thus, the Rigs from TEs can suppress or enhance gene expression including TE its own in transcription including the epigene and in post-transcription.
In plant, transitivity was found and miRNA-mediated cleavage of an RNA by transitivity can trigger the production of secondary siRNA [11,12]. The miRNA-dependent transacting siRNA (tasiRNA) as Rigs from the transitivity are generated from noncoding retrotransposons as well as protein-coding mRNA [13]. The secondary siRNA silenced additional genes. As this amplification of transitivity presents in human cells [14], we could expect miRNA* from the transposons to promote secondary siRNA production on its own targets, such as own retrotranspson’s transcript and host pri-miRNA [1]. Recently the miRNA gene has been reported to control its own pri-miRNA in the nucleus [15]. Although the endogenous siRNA and piRNA have already been reported to be involved in epigenetic transcriptional suppression [16,17], it is possible that the Rigs derived from the retrotransposon inactivate the neighboring and viral protein gene expression, while HIV-1 siRNAs affect gene expression of infected host cell [18] and tasiRNA-like offset miRNAs (moRNAs) are involved in the epigenetic regulation of gene expression in human cell line [19]. Further, some plantsor food-derived miRNAs accumulate in the serum of humans and could regulate gene expression of food-derived miRNA incorporated individuals (Figure 1(a)) [20]. These data suggest that sequence specific information of mobile Rigs could be available for fine-tuning of gene expression among the kingdoms of plants, animals and humans via foods and/or viruses. Since a part of sequences in human immunodficiency virus type 1 (HIV-1) miR-N367 is conserved in the plant miRNA, such as osa-miR-394 [21], one of LTR transposon HIV-1 may be transmitted from plant foods to chimpanzee, and then humans.
Edible vaccines or plant immune vaccines have been investigated as cost-effective ones against infectious diseases including human viral infections, such as human papilloma virus (HPV), hepatitis C virus (HCV), hepatictis B virus (HBV), rabis virus, Epstein-Barr virus (EBV) and human immunodeficiency virus (HIV) [22-29]. Oral vaccines are desirable from several stand points; 1) plant vaccines can serve low cost; 2) they are not accompanied by contamination due to endotoxins or pyrogens during purification of vaccines; 3) the cold chain for maintenance and trained medical staff for injectable vaccines/ sterile needles are not needed in the developing countries as well as developed countries; 4) They have lots of ecological parameters. Although the plant vaccines were developed in more potent immunogen and they were evaluated with immunogenicity, at that time, there was no idea that Rigs with the edible immunogens could be incorporated into the human cells and the edible Rig agents as a vaccine also keep above advantageous points. The edible Rig and anti-Rig vaccine, which could be derived from
 (a)
(a) (b)
(b)
Figure 1. Mobile Rigs in foods. (a) Sequence specific information of mobile Rigs could be available for fine-tuning of gene expression from the kingdoms of plants, animals to humans via foods with nutrients. Plant MIR-156a and MIR-168a have been incorporated into intestinal cells and transported circulating system, and then both plant miRNAs affect human metabolisms. These xenotropic miRNA would be retrohomed into the human genome because the human genome involves orthologs of plant miRNAs; (b) Environmental Rigs were contained in foods (plants and meat), viruses and others as environmental quantum energy including X-ray. The RNA wave model 2000 is; 1) the Rigs as a mobile genetic element induce transcriptional and post-transcriptional silencing via networking-architecture; 2) the Rigs expand into the environmental cycle of life; 3) the Rigs can self-proliferate; 4) the Rigs have two types of information as resident and genomic ones. This model is based on superposing quantum states of each other’s Rigs from natural environments, such as the Japanese Shrine woods (as its setting) according to Schrödinger’s “What is life?” [138,139]. Therefore, the food/virus-derived Rigs and chemicals with nutrients were fed/infected into humans and accumulate in the serum of feeding/infected humans, and then the Rigs as mobile ones could regulate human gene expression in a quantum RNA language (QRL)-specific manner, Further, the environmental stress, such as temperature and UV based on quantum energy could affect the quantum states of QRL in humans. Both cases would occasionally induce several Rig diseases (RigDs), such as autoimmune diseases, heart failure, hepatic disorder, neurodegeneration, neoangiogenesis, metabolic diseases, DNA gene diseases, viral infection, inflammation, cancers, fibrosis, hematopoietic diseases, atherosclerosis and cardiovascular diseases, etc.
Rig as well as its derivatives-expressing or Rig-contained foods, could directly control the life tuning and would be evaluated with Rig profiling.
The environmental Rigs are in foods, viruses and others, such as humans, Japanese shrine woods and dust in nature and could induce several Rig diseases (RigDs) with environmental stresses and chemicals (Figure 1(b)), therefore, the environmental stresses can cause alter of the Rig profiles [30]. Further, the mobile miRNA genes could be delivered by RNA gene vectors, foods as well as stem cells, therefore, in turn, all gene therapies, stem cell therapies and therapies with diets are the vehicular system of delivery for the Rigs. The highly active antiretroviral therapy (HAART) could efficiently repress HIV- 1 replication to the viral level of low or undetectable in the HIV-1-infected individuals [31]. But survivors remained as viral reserved patients with long-term complicated diseases, such as cardiovascular disease, liver and renal failure, neurodegeneration and malignancy [32-36] as less achievement of reconstitution in complete CD4+ T cell numbers [37,38]. Therefore, autologous and allogeneic hematopoietic stem cell transplantation have been performed to cure long-lived viral reservoirs [39]; however, no clinical benefit or transient increased CD4 counts has been mainly observed by lots of reports [40]. Although allogeneic CCR5∆32 homozygous T cell transplant together with myeloablative therapy can suppress HIV-1 replication and eradicated HIV-1 reservoirs upon only one clinical outcome [41], the result from the allogenic stem cell therapy cannot have yet been clearly explained the reason why HIV-1 was eliminated. Further, rare CCR5∆32 donors for stem cell therapy were found in the geographical search [42]. Recently, allogeneic HIV- 1-resistant CCR5∆32 donors T cells with ZFN gene transduction have been planed to be used for stem cell therapy and the first phase I clinical trial is being evaluated with CCR5 KO T cells (clinicaltrials.gov NCT00842634 in US), but it is not autologous stem cells because infected patients’ autologous stem cells or induced pluripotent stem (iPS) cells cannot used for the stem cell research to be safe; together with lots of failures of stem cell therapy for HIV-1 infection with autologous T cells [40]. Further, although artificially reprogrammed allogeneic cells from CCR5∆32 donors T cells would be used for HIV-1 infection, the endpoint of therapy with the iPS cells would be faced on xenotropic microRNA problem [43]. Thus, in HIV-1 stem cell therapy, there is no idea that mobile Rigs from the implanted stem cells could have a responsible for HIV-1 replication. The mobile Rigs for HIV-1 therapy are completely dropped in all papers about HIV-1 eradication because nobody investigated foods which above patient ate. In this paper, we would explain about the possible Rig vaccines for HIV-1 eradication in the longitudinal HIV-1 infection and into perspective.
2. Genetic Information of miRNAs
2.1. miRNAs and TEs
miRNAs are the dominance of small endogenous RNA, approximate 20 nucleotides (nts) in humans and the miRNA genes are produced from non-coding regions and coding regions containing the introns or exons in the human genome [44]. These endogenous small RNAs can inhibit or augment posttranscriptional events and control transcription [45,46]. The endogenous short interfering RNA (end-siRNA) and miRNA genes interact with argonaute (Ago) protein and small RNA/Ago complex leads to translational suppression [47,48] or mRNA decay [49] and the Piwi subfamily of Ago proteins would be restricted to express in the germ cells but miRNAs inhibit the expression of TEs in the genome of germ cells [50]. Subsequently, small RNAs could regulate gene expression and retroelement activation. It suggests that functioning of some miRNAs may be Ago-independent about TE regulation. TE has been major producers of the small RNA as described above; therefore, the retroposable Rigs could be retrohoming to germ line haploid via blood circulation, then sperms and eggs could carry the resident miRNAs and during stem cell development, the small RNAs could act to maintain the TE in a silenced state [51-53]. Further, in animal epigenome, mouse TEderived piRNAs guide DNA methylation to TEs [17] and small RNAs also cross-talk heterochromatin modification [16]. Thus, it is simply shown that the small RNA is always required for the regulation of TEs [54], even if HIV-1 infection because HIV-1 RNA would be processed to small RNAs in the cytoplasm of cells [55,56]. On the next subject, the switch on-off mechanism among miRNA/mRNA RNA interactions are discussed for the control of TEs.
2.2. The Seed Theory with miRNAs
In the case of translational silencing, the 5’ end of and the 3’ end of small RNA bind to the MID domain and the PAZ domain of Ago, respectively [48]. The seed (nucleotides 2 - 7) of miRNA targeted to >90% of the 3’UTRs of the human genome [57-59]. The seed region of miRNA/Ago can target to the 3’ untranslated region of mRNAs and the GW protein family also binds to Ago, then the GW induces deadenylation of the target mRNA by recruiting deadenylases. Further, poly(A) of mRNA is also shortened or mRNA is decapped by mRNA decay system [46]. However, recently, translational suppression of miR-430 has been happened before initiation phase and before mRNA decay of the translation pathway in zebrafish [60] or in Drosophila, therefore, miRNA-mediated silencing inhibited translation at an early step in vivo [49]. The explanation of miRNA-dependent translational suppression is always one to one system; however, the seed of miRNA and mRNA are not one to one interaction [61] and actually mRNAs targeted by more than two miRNA seeds have increased expression variability in human brain [62] and the seed less miRNA inhibited target mRNA translation [63]. Food-derived mature plant MIR-168a targets exon 4 of LDLRAP1 protein coding region and decreased the protein level but did not affect the mRNA level in vivo [64] and Ago/mRNA binding can take place in the absent of miRNA [65]; suggesting that translational suppression mechanisms by small RNAs is not so simple ones and miRNA is information but not the system. The system of the Ago would be analogous to a Turing machine of the quantum computer [66], and miRNA is income of information and DNA is its hard copy, therefore, it is assumed that the miRNA genes are the language, which is the expression of life.
2.3. RNA Wave Model with the Quantum RNA Language (QRL)
Although the proposition that life is fine-tuned by miRNA information is generally accepted [67], the mechanisms of alteration of the miRNA profile by the environmental factors without Rigs remain elusive. The RNA wave model 2000 consists, 1) the Rigs as a mobile genetic element induce transcriptional and posttranscriptional silencing via net working-processes; 2) the RNA information is supplied by the Rigs expands to intracellular, intercellular, intraorgan, interorgan, intraspecies, and interspecies under the cycle of life into the global environment; 3) the mobile Rigs can self-proliferate; and 4) cells contain two types information as resident and genomic Rigs [66]. Based on the RNA wave, the mobile miRNA genes are information but not the system [67,68], therefore, the Rigs can fine-tune our life, and evolution, development, proliferation, apoptosis, oncogenesis, signaltransduction, immune-reaction, metabolism, mutation and recombination of the human genomic DNA in somatic and germ cells, and behaviouring [67]. It means that Rigs are involved in most of all events in life tuning as causes, results and processes. Dysregulation and mull-function of the Rigs are involved in the pathogenesis of most of all human diseases, cancer, neurodegeneration, heartfailure, infectious diseases, inflammation, fibrosis, atherosclerosis, cardiovascular diseases, abnormal behaviour, metabolic diseases, etc. (Figure 1(b)). In the case of cancer, it is well known that the miRNA profiles in the tumor cells are changed and magnetic resonance image (MRI) is used for detection of tumor parts in organs; therefore, miRNA electron spin should be changed. The pulse waves of the electrons in the organ hydrogen atoms of water in the MRI could be derived from a source of the magnetic electron spin produced by altered resident miRNA electron spin. It is actually shown that alteration of electron spin by sensitive spin-labeling electron spin resonance (ESR) was detected in miR-125b expressed breast cancer cells when compared with less miR-125b expressed control cells [69]. Thus, the superposing of the electron spin direction in the high dimensions of Rigs has been introduced as the quantum RNA language (QRL) [67].
2.3.1. The Limits of the Seed Theory
The miRNA genes’ number has been reported to be 202,765 per a cell and Ago 1 to 4 molecules were approximately 15,000 - 17,000 [70,71]. Therefore, >90% of the miRNA genes did not bind Ago proteins, further, mRNAs would be associated with a seven-fold excess of miRNA from Janas et al. data. Their data suggest that it does not make sense about the stoichimetric model of miRNA-mediated repression for the gene expression and they speculated that Ago supports a model of catalytic function of Ago with P-body-independent in translational repression, that contains that Ago-free miRNAs are stabilized by binding to target mRNAs, and target mRNAs are destabilized when their miRNA recognition elements are occupied by miRNAs, allowing transient Ago binding. Although the average G/C content of seed sequences was higher than that of mature miRNA sequences, suggesting that an association between the degree of functionality of the sequence and its average G/C content [72], they did not find statistically significant differences in the ability of seedless and seed-region-containing miRNAmRNA duplexes to be repressed. Furthermore, there was no discussion about how Ago-free miRNAs are stabilized and target mRNAs are destabilized catalytically because of Ago enzyme-free. The data suggest that the thermodynamic stability of seed-dependent miRNA-target mRNA duplexes is not completely involved in mechanisms upon translational repression of gene expression. When target mRNA has been replaced with target TE, amounts of total Ago protein would be shortened more. And although CeRNA hypothesis explained that the miRNAmRNA interaction itself is a language of RNA [73], in the case of the long non-coding RNA such as TE, it is not yet known whether Ago system and base-pairing system have any responsible for suppression system of TE expression or not [74].
2.3.2. The QRLs
Before the seed theory has been established, we have developed in new algorithms for the miRNA genes using quantum bits as physicochemical characters of RNA bases with measurement of electron spin according to above RNA wave model [67,75] (see Figure 2). In the Rig quantum code with the fragment molecular orbit method (FMO), positive potencies of the G base cluster
 (a)
(a)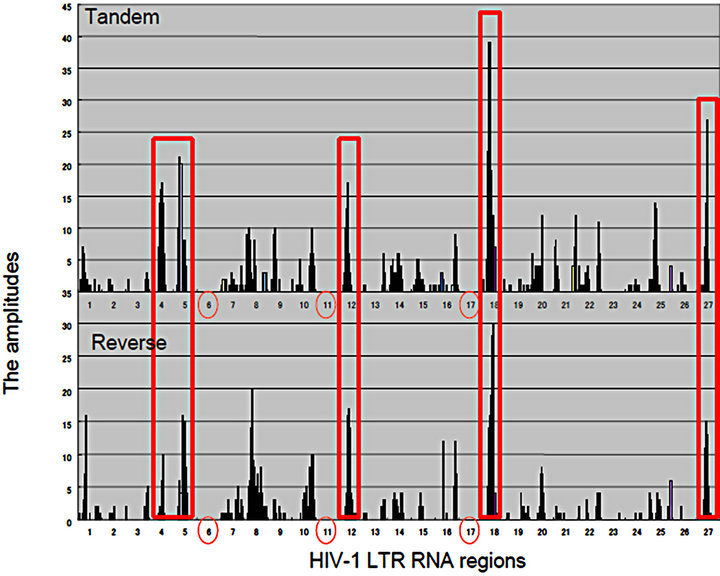 (b)
(b)
Figure 2. Matrics with Rigs. (a) RNA wave-based gene regulation was investigated in HCV infection on the quantum inner space. The fragment molecular orbit methods (FMO)-derived values of RNA bases were calculated by winMOPAC software version 3 and binary transformation of each base was performed as described previously [67]. The superposing state of QRLs of HCV site 2 (|f(x)>) and miR-122 (|x>) was applied for the matrix of 5’ → 3’ tandem direction and 3’ → 5’ reverse orientation. The values of superposed samples were shown as amplitude vectors of an intersecting point between miR-122 and HCV site 2 ket modules in the matrix as the 2 quantum bits. The HCV genotypes, 1a, 1b, 2a, 3a, 4a, 5a and 6f were analyzed in the matrix. The amplitude values of intersecting points in matrices were made with histogram by Excel platform [140]. The 5’ → 3’ tandem direction and 3’ → 5’ reverse orientation of miR-122 were represented in the left and right panels, respectively. The seed regions of miR-122 were shown as green colored squares; (b) The HIV-1 LTR was divided into 27 regions, and the 5’ → 3’ direction of the LTR regions. The amplitude vectors (2y) in each region were made with histogram by Excel platform and were represented with histograms. Three entangled QRLs, LTR plus miR-N367, LTR plus miR-#4 and LTR plus miR-H1 were applied for a matrix of 5’ → 3’ tandem direction and the 3’ → 5’ reverse orientation. The RNA regions, 6, 11 and 17 containing the RNA chain of viral miRNA were emptied (red circles). The cut off value of 16.0 at the mean amplitude was given by the red line in the histogram. TAR: Tat-responsive region. Comparisons between the 5’ → 3’ tandem direction matrix (the upper panel) and the 3’ → 5’ reverse orientation one (the lower panel). Both histograms were represented and the high amplitude values in the region 4, 5, 12, 18 and 27, BE condensation (BEC)-like were enclosed with red squares.
was found. Although there is a hypothesis that the G and C bases would expand toward the C, G, A and U ones in the early evolution of RNA [76,77], canonical RNA editing occurs in A-to-I and C-to-U of 3’ UTRs at miRNA target sites [78]; therefore, the G base is stable to the RNA editing although the I base is recognized as the G base. Recently, the seed-related G-rich motif has modulated miRNA-directed host mRNA regulation in mouse embryonic stem (ES) cells [79] and mRNAs harboring G-bulge sites were bound by the seed of miR-124 in mouse brain [80]. Therefore, these results suggest that the G base is the important pivot nucleotide for transmission of RNA information. However, there is a long seed through whole sequences of miRNAs and long noncoding RNA (lncRNA) can control gene translation in brain and hematopoietic cells [81-83]. Further, it has been reported that seed matches are not a sufficient predictor for C. elegans lsy-6/the cog-1 homeobox gene mRNA 3’UTR interaction [84] and in zebrafish, miR- 214 without canonical seed paring can effectively target a mRNA for silencing [63]. Relation between HCV site 2 RNA and the seed sequence of miR-122 has been reported that the 3’ tail nucleotide miR-122 is important for recognition of HCV site 2 in selective 2’-hydroxyl acylation analyzed by primer extension (SHAPE) chemistry assay [85] and this kind of 3’ tail effect has previously been reported [58, 86]. These data suggested that the seed pairing of the 5’ → 3’ to the 3’ → 5’ nucleotide (nt) sequences is not enough to explain how Rigs to develop bioinformation. The superposing quantum states of Rigs with a quantum-based algorithm were preliminarily shown in Figure 2, between genotypes of hepatitis C virus (HCV) site 2 and miR-122 by QRL (Figure 2(a)). The QRL was independent of the seed region in miR-122. Further, when the entangling quantum phase between HIV-1 3’LTR and miR-N367, #4, and H1 was investigated by the QRL, the QRL was also independent of the seed region in miRNAs (Figure 2(b)). When compared quantum state of the entangled three miRNAs/the HIV-1 LTR, this case showed condensation of spin, like spin squeezing in a Bose-Einstein condensates (BEC) (red squares: Figure 2(b)) [87,88]. Since there are not ultracold atoms, the condensation of miRNA electron spin may display quantum behaviour and function of miRNA such as the QRL and the altered miRNA BEC in the cells may dominantly affect the pulse waves of the electrons in the organ hydrogen atoms of water in the MRI.
2.4. HIV-1 as a Rig
While de novo activation of TEs causes genetic disorders [4,89] and somatic retrotransposition of TE has been reported to be active in the human brain [90], except for the epigeneitic regulation of TEs, there is less explanation for the control mechanisms of the observed transposon activity. We have shown that one of TEs, HIV-1 provirus in the human genome can be regulated by miRNA, miR-N367 in transcription and translation [91, 92] and Chen et al. [53] have reported that endogenous siRNAs, endo453 and endo392 can silence human long interspersed nuclear element 1 (LINE-1) activity through DNA methylation. Further, the ribonuclease (RNase) III enzyme Drosha can process virus-derived cytoplasmic primary miRNA in the cytoplasm [55]. These data suggest that the resident miRNAs are localized in the cytoplasm, therefore, transcripts of the TE could be processed to miRNAs after a transfer of the transcripts in cell-tocell communication through the microsomes [55,93,94] and crossing inter-kingdom boundaries through the diet [20,21,75,95]. About criteria 3 of the RNA wave; the Rigs can self-proliferate. At first step, transitivity was found as described above that miRNA-mediated cleavage of an RNA can trigger the production of secondary siRNA in the human cells [14]. Further, RNA-directed RNA polymerase (RdRP)-like proteins and an RdRP ribozyme C47U were identified [92,96-98], suggesting that both host and donor Rigs could be amplified in the cytoplasm of human cells. Even if in the case of DNA viruses, EBV virions contain viral mRNAs, miRNAs, and other noncoding RNAs and these RNAs controlled early steps of EBV infection to B cells [99], therefore, robustness of the host human genome may be affected by the interaction between these mobile Rigs and the host ones. Thus, since the superposing state of HIV-1 LTR and miR-N367, #4, and H1 was shown in QRLs in Figure 2(b), that of QRLs would initiate the latency of HIV-1 and the aberration of miRNA BEC by environmental stresses may induce HIV-1 reactivation. Further, new avenues may open the door to investigate superposing between the mobile Rigs and the host ones for life tuning under quantum computing.
2.5. Mobile Rigs
Based on above RNA wave concepts, cellular, the individual and species’ communication in humans could have been mediated by the transmission of the resident Rigs in extracellular microvesicles, such as ectosome (approximate 100 nm - 1 µm in diameter) or exosomes (approximate 50 - 100 nm), and in vesicle-independent form, such as high-density lipoproteins (HDLs) [93,94]. The exosomerelated miRNA secretion has been reported to be implicated in ceramide dependent manner [100] and the packaging of miRNA into the exosomes was selective [101]. Further, the non-vesicular and vesicular secretion of miRNAs were associated with Ago2 [102-104]. Although miRNAs are present in various body fluids and could be selectively included into above exosomes, like retroviral particles, recent researches unveiled that breast milk contains miRNAs in exosomes [105,106]. These milk exosomal miRNAs can be orally delivered to the reciepient cell of the infant offspring with nutrients and could control brain development, growth, and healthy immune system, further, may transmit acquired inheritable phenotype of donors [43]. From this aspect, since exogenous retroviruses and endogenous TEs use micro vesicle pathway elements, such as exosomes [107], HTLV-1 can orally transmit from mother to infant via breast milk vertically and induces human malignancy [108,109]. These results suggest that the Rigs is the mobile genetic element via food, human has used the food-derived Rigs for maintenance of homeostasis and incorporated them into the genome for evolution.
3. The miRNA Gene for the HIV-1 Vaccine
HIV-1 replication is regulated by the viral and cellular miRNA genes. Anti-HIV-1 drugs inhibit reversetranscription (zidovudine, didanosine, zalcitabine, lamivudine, efavirenz, nevirapine, delavirdine mesilate, sanilvudine, emtricitabine and tenefovir), integration (raltegravir, elvitegravir) and viral protease (saqunavir mesilate, saquinavir, ritonavir, lopinavir, indinavir sulfate ethanolate, nelfinavir mesilate and amprenavir) activities for virus particle proliferation. Further, by truvada (emtricitabine and tenefovir) the regimen of HIV-1 infection was simply progressed from high active anti-retrovirus therapy (HAART) and stribild (emtricitabine, tenefovir and elvitegravir) would go on as a preventing agent of HIV-1 infection (Figure 3(a)). However, there is no HIV-1 inhibitor agent for transcription of viral mRNA and viral genome RNA from provirus, and translation of viral proteins for viral particle components. And these chemotherapeutic agents could make drug-resistant HIV-1 clones. Recently, the implication of miRNA and HIV-1 has been reported by several research groups because of its complexes [110- 116]. The different virus clones and infecting target cells showed different expression of the miRNA genes. Further, small viral RNAs of approximate 26,000 sequences
(18 - 22 nts) have been detected in HIV-1 infected SupT1 cells by deep sequencing [115]. Viral siRNAs, such as vsiR-298, vsiR-384, vsiR-3228, vsiR-6496, vsiR-7341, vsiR-8200, vsiR-8943, and vsiR-9095 can target sense HIV-1 mRNAs [115]. On the other hand, viral miRNAs, miR-H1, miR-N367, miR-TAR-5p and miR-TAR-3p target the HIV-1 nef/LTR sequences, and cellular hsamiRNAs, miR-29a, miR-28, miR-125b, miR-150, miR- 223, miR-382, miR-133b, miR-138, miR-149 and miR- 326 also target the nef/3’ LTR sequences [117,118]. Furthermore, expression of HIV-1 transcription positive factor, PCAF, Cyclin T1 and Pur-α were suppressed by miR-17-5p and miR-20a, miR-198, miR-27b, miR-29b, miR-150 and miR-223, and miR-15a, miR-15b, miR-16, miR-20a, miR-93 and miR-106b, respectively [117,118].
These viral and cellular anti-HIV-1 promoter miRNAs decreased viral replication (Figure 3(b)), suggesting that Rigs could have responsible for the latent state of HIV-1 and promoter region-related mi-RNAs could be effective repressors of HIV-1 replication. On possible idea is that mixed cocktails of miRNAs for HIV-1 infection may be possible to use as HIV-1 edible vaccines [75,92].
 (a)
(a) (b)
(b)
Figure 3. Anti-HIV-1 drugs and miRNAs in the viral life cycle. (a) No HIV-1 inhibitors for viral transcription and translation. Viral attachment via CD4 and CXCR4 and/or CCR5 receptor is inhibited by virus binding inhibitor. After viral entry and uncoating, reversetranscription is inhibited by reversetranscriptase inhibitor. Double stranded viral DNA is transported to the cell nucleus and then integrated into the genome by viral integrase, which can be blocked by integrase inhibitor. From the provirus in the genome, viral genomic RNA and viral proteins are expressed but there is no inhibiting agent, therefore, miRNA or anti-miRNA agents may be available for HIV-1 eradication. Viral assembly and budding occur and progeny virion maturation is inhibited by protease inhibitor; (b) Anti-HIV-1 cellular miRNAs. Viral post-transcriptional events are inhibited by cellular miRNAs directly or indirectly via cyclin T1, PCAF and Pur-α.
4. An Anti-miRNA Agent, Miravirsen
MiR-132 enhances HIV-1 replication, therefore, antimiRNA oligonucleotide (AMO) would be applicable for therapy HIV-1 infection [119]. As concerning to longitudinal HIV infection, treatment for dual and single chronic HBV and/or HCV co-infection have to set up because of 3% - 5% of HIV-infected individuals [120]. MiR-122 is the most dominant miRNA gene (approximate 70% of the total miRNA) expressed in the liver. The miRNA gene can control lipid and cholesterol metabolism and maintain the levels of the plasma cholesterol [121]. Although lipid and cholesterol metabolism are regulated on day-light dependence, miR-122 targeted the circadian rhythm of the metabolic regulation in the liver [122]. Further, mouse miR-122 knockout (KO) in germline or in liver-specific resulted in hepatosteatosis, hepatitis, and the development of tumor, therefore, miR-122 functioned as tumor suppressor [123]. The expression of miR-122 in the liver permits HCV 1b replication [124]. And the HCV replication was repressed about 80% by anti-miR- 122. The miR-122 gene can bind to the 5’UTR of HCV genomic RNA [125] and stimulate HCV protein translation [126] but contradictorily, miR-122 did not directly stimulate HCV replication [127,128]. Although treatment of anti-miR-122 LNA, miravirsen reduced HCV RNA in serum of HCV-infected individuals on Phase IIa trial [129], in the case of chronic hepatitis C, there is no correlation between miR-122 and HCV RNA levels of patients [130]. The AMO LNA is clearly effective to HCV infection but information of miR-122 to HCV life cycle has not yet been ciphered. On the contrary, miR-122 has been reported to be suppressed in chronic HBV infected patient [131] and the reducing expression of miR-122 in chronic HBV was explained through expression inhibition of HBV enhancer HNF3 and HNF4α because miR- 122 requires HNF for its own transcription [132]. The miR-122 mimic LNA may be considered as a candidate of HBV therapy. For HIV-1-infected individuals, miR- 122 was increased in HIV-1-infected T cells [133]. In a scenario, if HIV-1 replication could be augmented in CD4+ T cells and macrophages by miR-122, it is plausible whether miravirsen can be examined for HIV-1 infection as well as dual HIV infection with HCV or not because the PBMCs pool of the liver. At a computation analysis, miR-3065-3p has recently been predicted as an candidate of antiviral therapeutic agent for HIV-1, HCV and HBV triple infection [134]. We have to challenge ahead to cure the mixed infection for HIV-1 eradication.
5. Linking Rig’s Pieces of Edible Vaccine from Stem Cell Therapy for HIV-1
Corresponding to the evidence-based medicine, it took more than 25 years to treat infection of HIV-1 with allogenic hematopoietic cell transplantation therapy using stem cells derived from bone marrow or peripheral blood; however, the evidence is that most of all clinical trials were unsuccessful [40]. Since use of treatment of HAART, the opportunity infections were decreased and the mortality among HIV-1 infected individuals increased. However, the incidence of malignancies, such as nonHodgkin’s lymphoma arises in HIV-1 infection [35,36,38] and the lymphoma has still been remained as the most lethal complication of AIDS. Recently a HIV-1 patient arisen acute myeloid leukemia (AML) has been reported to success the reconstitution of CD4+ T cells and reducing of the size of the potential HIV-1 reservoir after stem cell therapy with CCR5∆32 donors [41]. Consequently, the patient remains without any evidence of HIV-1 infection for more than 3.5 years after discontinuation of anti-retroviral therapy. However, before the stem cell therapy, the patient was treated with cyclophosphamide and a 400 or 200-cGy total body irradiation as immunosuppressive treatment with anti-lymphoma antibodies, gemtuzumab, and anti-lymphoma agents, amsacrine, fludarabine and cytarabine at first transplantation and the second one. Further, CXCR4 expression was normal on recovered CD4+ T cells, but CXCR4 susceptible HIV-1 also be not detected, therefore, X4 HIV-1 did not rebound. Although the point of HIV-1 eradication was not so clear, there is an unconvincing evidence that CCR5- ∆32 stem cell therapy was effectively replaced with donor-derived CD4+ T cells and HIV-1 was eradicated. Even if there were CCR5 deleted stem cells or non-tumorigenic and complete reprogrammed iPS cells and the cells would be used for stem cell therapy [42], the allogenic transplantation would have the similarly unsure problems, such as the low efficacy, graft versus host (GVH) reactions, and harmful interaction between antiHIV drugs and anti-GVH agents. Further, on early time, CCR5∆32 heterozygosity has not been observed to inhibit HIV-1 transmission in homosexual or hemophiliac populations [135] and in uninfected individuals with a range of HIV-1 exposures, the prevalence of homozygosity for the CCR5∆32 allele increases with increasing HIV-1 exposure [136] because several non-CCR5 receptors, such as CCR3 and CCR2b are HIV-1 entry cofactors. Therefore, non-X4 HIV-1 might be transmissible in individuals CCR5∆32 homozygous and CCR5∆32 is not an absolute protection factor. In addition, while the miRNA genes are not recognized for the stem cell therapy at all, the relations between miRNAs and lymphoma or between miRNAs and stem cell therapy, or between miRNAs and chemotherapy in the HIV-1 patient are in the cloud. Recently, clinical transplantation of the tracheobronchial airway with a stem cell has been reported and miRNA expression of patients were monitored [137]. If we could investigate miRNA gene profiles of a HIV-1

Figure 4. Application of sequence specific QRL for human diseases. The QRL may operate not only fine tuning of gene expression but also the programmed evolution, therefore the QRL could be related with spontaneous RigDs and inheritable ones. The information technology-based therapy (iTBT) might be involved into quite near future progress of therapy and prevention for RigDs, such as HCV (the middle panel) and HIV-1 infections (the right panel). The left panel represented that Rigs in foods could be affects our health and metabolisms. The middle panel showed that miravirsen can prevent HCV proliferation. The right panel represented that Rigs in stem cell implantation would be produced from allogeneic stem cells and may affect the HIV-1 infected patients as the first previous description about the application of the miRNA gene for HIV-1 prevention [141]. Thus, the concept of the QRL is common to the whole panel and may be applied for eradication of HIV-1 as the medical and manufacturing new devices with chemotherapy and stem cell therapy.
infected patient for allogenic CCR5∆32 homozygous T cell transplant from CCR5∆32 donors, we might elucidate the question which miRNAs and QRLs can suppress HIV-1 replication and eradicate HIV-1 reservoirs. We know the host miRNA genes which can suppress HIV-1 replication described above, that may be an idea for therapeutic application for HIV-1 eradication, such as the edible Rig vaccine. Subsequently it would be concluded that therapy for lymphoma and eradication of HIV-1 are quite differently biological challenges; however, as described in miR-122 about HCV replication and tumor suppressor, a Rig may be implicated in both viral pathogenesis in the longitude alive HIV-1 patients and AML tumorigenesis. Further, as shown in Figure 4, xenotropic miRNAs in foods and miravirsen were analogous on the concept of principle under the QRL as the information transmission. If xenotropic miRNAs in microvesicles were actively released from allogenic CCR5∆32 stem cells, cure effect of CCR5∆32 homozygous T cell transplant may be caused by the xenotropic Rigs from the transplant cells (Figure 4), furthermore, its efficacy may be implicated in the dilution effect. Since allogenic CCR5∆32 stem cell therapy should not be valued without Rig idea in the lineage specificity even if iPS cells could be used, pharmacological approach with Rigs would be more relevant for HIV-1 eradication than stem cell therapy. Furthermore, the global scale of the HIV-1 epidemic needs to cure large numbers of patients in the limited settings. Thus, miRNA assessment is the most important for challenging HIV-1 eradication and edible Rig vaccine as the QRL may be suitable for the longitude HIV-1 cure together with chemotherapy and stem cell therapy.
REFERENCES
- Y. R. Fujii, “RNA Genes: Retroelements and Virally Retroposable microRNAs in Human Embryonic Stem Cells,” The Open Virology Journal, Vol. 4, 2010, pp. 63-75. doi:10.2174/1874357901004010063
- H. H. Kazazian Jr., “Mobile Elements: Divers of Genome Evolution,” Science, Vol. 303, No. 5664, 2004, pp. 1626- 1632. doi:10.1126/science.1089670
- S. R. Wessler, “Transposable Elements and the Evolution of Eukaryotic Genomes,” Proceedings the National Academy of Sciences the USA, Vol. 103, No. 47, 2006, pp. 17600-17610. doi:10.1073/pnas.0607612103
- R. Cordaux and M. A. Batzer, “The Impact of Retrotransposons on Human Genome Evolution,” Nature Review Genetics, Vol. 10, No. 10, 2009, pp. 691-703. doi:10.1038/nrg2640
- J. Li, Y. Liu, X. Xin, T. S. Kim, E. A. Cabeza, J. Ren, R. Nielsen, J. L. Wrana and Z. Zhang, "Evidence for Positive Selection on a Number of microRNA Regulatory Interactions during Recent Human Evolution,” PLoS Genetics, Vol. 8, No. 3, 2012, Article ID: e1002578. doi:10.1371/journal.pgen.1002578
- N. V. Rozhkov, N. G. Schostak, E. S. Zelentsova, I. A. Yushenova, O. G. Zatsepina and M. B. Evgen’ev, “Evolution and Dynamics of Small RNA Response to a Retroelement Invasion in Drosophila,” Molocular Biology and Evolution, Vol. 30, No. 2, 2012, pp. 397-480. doi:10.1093/molbev/mss241
- C. Feschotte, N. Jiang and S. R. Wessler, “Plant Transposable Elements: Where Genetics Meets Genomics,” Nature Review Genetics, Vol. 3, No. 2, 2002, pp. 329-341. doi:10.1038/nrg793
- P. S. Schnable, D. Ware, R. S. Fulton, J. C. Stein, F. Wei, S. Pastemak, C. Liang, J. Zhang, L. Fulton, T. A. Graves, P. Minx, A. D. Reily, L. Courtney, S. S. Kruchowski, C. Tomlinson, C. Strong, K. Delehaunty, C. Fronick, B. Courtney, S. M. Rock, E. Belter, F. Du, K. Kim, R. M. Abbott, M. Cotton, A. Levy, P. Marchetto, K. Ochoa, S W. M. Jackson, B. Gillam, W. Chen, L. Yan, J. Higginbotham, M. Cardenas, J. Waligorski, E. Applebaum, L. Phelps, J. Falcone, K. Kanchi, T. Thane, A. Scimone, N. Thane, J. Henke, T. Wang, J. Ruppert, N. Shah, K. Rotter, J. Hodges, E. Ingenthron, M. Cordes, S. Kohlberg, J. Sgro, B. Delgado, K. Mead, A. Chinwalla, S. Leonard, K. Crouse, K. Collura, D. Kudrna, J. Currie, R. He, A. Angelova, S. Rajasekar, T. Mueller, R. Lomeli, G. Scara, A. Ko, K. Delaney, M. Wissotski, G. Lopez, D. Campos, M. Braidotti, E. Ashley, W. Golser, H. Kim, S. Lee, J. Lin, Z. Dujmic, W. Kim, J. Talag, A. Zuccolo, C. Fan, A. Sebastian, M. Kramer, L. Spiegel, L. Nascimento, T. Zutavern, B. Miller, C. Ambroise, S. Miller, W. Spooner, A. Narechania, L. Ren, S. Wei, S. Kumari, B. Faga, M. J. Lavy, L. McMaha, P. Van Buren, M. W. Vaughn, K. Ying, C. T. Yeh, S. J. Emrich, Y. Jia, A. Kalyanaraman, A. P. Hsia, W. B. Barbazuk, R. S. Baucom, T. P. Brutnell, N. C. Carpita, C. Chaparro, J. M. Chia, J. M. Deragon, J. C. Estill, Y. Fu, J. A. Jeddeloh, Y. Han, H. Lee, P. Li, D. R. Lisch, S. Liu, Z. Liu, D. H. Nagel, M. C. McCann, P. SanMiguel, A. M. Myers, D. Nettleton, J. Nguyen, B. W. Penning, L. Ponnala, K. L. Schneider, D. C. Schwartz, A. Sharma, C. Soderlund, N. M. Springer, Q. Sun, H. Wang, M. Waterman, R. Westerman, T. K. Wolfgruber, L. Yang, Y. Yu, L. Zhang, S. Zhou, Q. Zhu, J. L. Bennetzen, R. K. Dawe, J. Jiang, N. Jiang, G. G. Presting, S. R. Wessler, S. Aluru, R. A. Martienssen, S. W. Clifton, W. R. McCombie, R. A. Wing and R. K. Wilson, “The B73 Maize Genome: Complexity, Diversity, and Dynamics,” Science, Vol. 326, No. 5956, 2009, pp. 1112-1115. doi:10.1126/science.1178534
- E. S. Lander, L.M. Linton, B. Birren, C. Nusbaum, M. Zody, J. Baldwin, K. Devon, K. Dewar, M. Doyle, W. Fitzhugh, R. Funke, D. Gage, K. Harris, A. Headford, J. Howland, L. Kann, J. Lehoczky, R. LeVine, P. McEwan, K. McKeman, J. Meldrim, J. P. Mesirov, C. Miranda, W. Morris, J. Naylor, C. Raymond, M. Rosetti, R. Santos, A. Sheridan, C. Sougnez, N. Stange-Thomann, N. Stojanovic, A. Subramanian, D. Wyman, J. Rogers, J. Sulston, R. Ainscough, S. Beck, D. Bentley, J. Burton, C. Clee, N. Carter, A. Coulson, R. Daedman, P. Deloukas, A. Dunham, I. Dunham, R. Durbin, L. French, D. Grafham, S. Gregory, T. Hubbard, S. Humphray, A. Hunt, M. Jones, C. Lloyd, A. McMurray, L. Matthews, S. Mercer, S. Milne, J. C. Mullikin, A. Mungall, R. Plumb, M. Ross, R. Shownkeen, S. Sims, R. H. Waterston, R. K. Wilson, L. W. Hillier, J. D. McPherson, M. A. Marra, E. R. Mardis, L. A. Fulton, A. T. Chinwalla, K. H. Pepin, W. R. Gish, S. L. Chissoe, M. C. Wendl, K. D. Delehaunty, T. L. Miner, A. Delehaunty, J. B. Kramer, L. L. Cook, R. S. Fulton, D. L. Johnson, P. J. Minx, S. W. Clifton, T. Hawkins, E. Branscomb, P. Predki, P. Richardson, S. Wenning, T. Slezak, N. Doggett, J. F. Cheng, A. Olsen, S. Lucas, C. Elkin, E. Uberbacher, M. Frazier, R. A. Gibbs, D. M. Muzny, S. E. Scherer, J. B. Bouck, E. J. Sodergren, K. C. Worley, C. M. Rives, J. H. Gorrell, M. L. Metzker, S. L. Naylor, R. S. Kucherlapati, D. L. Nelson, G. M. Weinstock, Y. Sakaki, A. Fujiyama, M. Hattori, T. Yada, A. Toyoda, T. Itoh, C. Kawagoe, H. Watanabe, Y. Totoki, T. Taylor, J. Weissenbach, R. Heilig, W. Saurin, F. Artiguenave, P. Brottier, T. Bruls, E. Pelletier, C. Robert, P. Wincker, D. R. Smith, L. Doucette-Stamm, M. Rubenfield, K. Weinstock, H. M. Lee, J. Dubois, A. Rosenthal, M. Platzer, G. Nyakatura, S. Taudien, A. Rump, H. Yang, J. Yu, J. Wang, G. Huang, J. Gu, L. Hood, L. Rowen, A. Madan, S. Qin, R. W. Dvis, N. A. Federspiel, A. P. Abola, M. J. Proctor, R. M. Myers, J. Schmutz, M. Dickson, J. Grimwood, D. R. Cox, M. V. Olson, R. Kaul, C. Raymond, N. Shimizu, K. Kawasaki, S. Minoshima, G. A. Evans, M. Athanasiou, R. Schultz, B. A. Roe, F. Chen, H. Pan, J. Ramser, H. Lehrech, R. Reinhardt, W. R. McCombie, M. de la Bastide, N. Dedhia, H. Blöcker, K. Hornischer, G. Nordsiek, R. Agarwala, L. Aravind, J. A. Bailey, A. Bateman, S. Batzoglou, E. Birney, P. Bork, D. G. Brown, C. B. Burge, L. Cerutti, H. C. Chen, D. Church, M. Clamp, R. R. Copley, T. Doerks, S. R. Eddy, E. E. Eichler, T. S. Furey, J. Galagan, J. G. Gilbert, C. Harmon, Y. Hayashizaki, D. Hassler, H. Hermjakob, K. Hokamp, W. Jang, L. S. Johnson, T. A. Jones, S. Kasif, A. Kaspryzk, S. Kennedy, W. J. Kent, P. Kitts, E. V. Koonin, I. Krof, D. Kulp, D. Lancet, T. M. Lowe, A. McLysaght, T. Mikkelsen, J. V. Moran, N. Mulder, V. J. Pollara, C. P. Ponting, G. Schuler, J. Schultz, G. Slater, A. F. Smit, E. Stupka, J. Szustakowski, D. Thierry-Mieg, J. Thierry-Mieg, L. Wagner, J. Wallis, R. Wheeler, A. Williams, Y. I. Wolf, K. H. Wolfe, S. P. Yang, R. F. Yeh, F. Collins, M. S. Guyer, J. Peterson, A. Felsenfeld, K. A. Wetterstrand, A. Patrinos, M. J. Morgan, P. de Jong, J. J. Catanese, K. Osoegawa, H. Shizuya, S. Choi and Y. J. Chen, “Initial Sequencing and Analysis of the Human Genome,” Nature, Vol. 409, No. 6822, 2001, pp. 860-921. doi:10.1038/35057062
- H.-M. Byun, K. Heo, K. J. Mitchell and A. S. Yang, “Mono-Allelic Retrotransposon Insertion Addresses Epigenetic Transcriptional Repression in Human Genome,” Journal of Biomedical Science, Vol. 19, No. 1, 2012, p. 13. doi:10.1186/1423-0127-19-13
- C. Johnson, A. Kasprzewska, K. Tennessen, J. Fernandes, G.-L. Nan, V. Walbot, V. Sundaresan, V. Vance and L. H. Bowman, “Clusters and Superclusters of Phased Small RNAs in the Developing Inflorescence of Rice,” Genome Research, Vol. 19, No. 3, 2009, pp. 1429-1440. doi:10.1101/gr.089854.108
- P. A. Manavella, D. Koenig, and D. Weigel, “Plant Secondary siRNA Production Determined by microRNADuplex Structure,” Proceedings the National Academy of Sciences the USA, Vol. 109, No. 7, 2011, pp. 2461-2466. doi:10.1073/pnas.1200169109
- M. Yoshikawa, A. Peragine, M. Y. Park and R. S. Poethig, “A Pathway for the Biogenesis of Trans-Acting siRNAs in Arabidopsis,” Genes Development, Vol. 19, No. 18, 2005, pp. 2164-2175. doi:10.1101/gad.1352605
- D. Langenberger, C. Bermudez-Santana, J. Hertel, S. Hoffmann, P. Khaitovich and P. Stadler, “Evidence for Human microRNA-Offset RNAs in Small RNA Sequencing Data,” Bioinformatics, Vol. 25, No. 18, 2009, pp. 2298-2301. doi:10.1093/bioinformatics/btp419
- X. Chen, H. Liang, C.-Y. Zhang and K. Zen, “miRNA Regulates Noncoding RNA: A Noncanonical Function Model,” Trends in Biochemical Sciences, Vol. 37, No. 11, 2012, pp. 457-458. doi:10.1016/j.tibs.2012.08.005
- E. M. Jobe, A. L. McQuate and X. Zhao, “Crosstalk among Epigenetic Pathways Regulates Neurogenesis,” Frontiers Neurogenesis, Vol. 6, 2012, p. 59.
- A. A. Aravin, R. Sachidanandam, D. Bourc’his, C. Schaefer, D. Pezic, K. F. Toth, T. Bestor and G. J. Hannon, “A piRNA Pathway Primed by Individual Transposons Is Linked to de Novo DNA Methylation in Mouse,” Molecluar Cell, Vol. 31, No. 6, 2008, pp. 785-799. doi:10.1016/j.molcel.2008.09.003
- N. C. Schopman, M. Willemsen, Y. P. Liu, T. Bradley, A. van Kampen, F. Baas, B. Berkhout and J. Haasnoot, “Deep Sequencing of Virus-Infected Cells Reveals HIV-Encoded Small RNAs,” Nucleic Acids Research, Vol. 40, No. 1, 2011, pp. 414-427. doi:10.1093/nar/gkr719
- R. J. Taft, C. Simons, S. Nahkuri, H. Oey, D. Korbie, T. R. Mercer, J. Holst, W. Ritchie, J. J.-L. Wong, J. E. Rasko, D. S. Rokhsar, B. M. Degnan and J. S. Mattick, “Nuclear-Localized Tiny RNAs Are Associated with Transcription Initiation and Splice Sites in Metazoans,” Nature Structural and Molecular Biology, Vol. 17, No. 8, 2010, pp. 1030-1035. doi:10.1038/nsmb.1841
- M. Jiang, X. Sang and Z. Hong, “Beyond Nutrients: FoodDerived microRNAs Provide Cross-Kingdom Regulation,” Bioessays, Vol. 34, No. 4, 2012, pp. 280-284. doi:10.1002/bies.201100181
- Y. R. Fujii and N. K. Saksena, “Viral Infection-Related microRNAs in Viral and Host Genomic Evolution,” In: K. V. Morris, Ed., RNA and the Regulation of Gene Expression, Horizon Scientific Press, London, 2008, pp. 91-107.
- P. Lenzi, N. Scotti, F. Alagna, M. L. Tornesello, A. Pompa, A. Vitale, A. de Stradis, L. Monti, S. Grillo, F. M. Bounaguro, P. Maliga and T. Cardi, “Translational Fusion of Chloroplast-Expressed Human Papillomavirus Type 16 L1 Capsid Protein Enhances Antigen Accumulation in Transplastomic Tobacco,” Transgenic Research, Vol. 17, No. 6, 2008, pp. 1091-1102. doi:10.1007/s11248-008-9186-3
- A. Fernandez-San Millan, S. M. Ortigosa, S. HervasStubbs, P. Corral-Martinez, J. M. Segui-Simarro, J. Gaetan, P. Coursaget and J. Veramendi, “Human Papillomavirus L1 Protein Expressed in Tobacco Chloroplasts SelfAssembles into Virus-Like Particles That Are Highly Immunogenic,” Plant Biotechnology Journal, Vol. 6, No. 5, 2008, pp. 427-441. doi:10.1111/j.1467-7652.2008.00338.x
- P. Madesis, M. Osathanunkul, U. Georgopoulou, M. F. Gisby, E. A. Mudd, I. Nianiou, P. Tsitoura, P. Mavromara, A. Tsaftaris and A. Daya, “A Hepatitis C Virus Core Polypeptide Expressed in Chloroplasts Detects Anti-Core Antibodies in Infected Human Sera,” Journal of Biotechnology, Vol. 145, No. 4, 2010, pp. 377-386. doi:10.1016/j.jbiotec.2009.12.001
- A. Modelska, B. Dietzschold, N. Sleysh, Z. F. Fu, K. Steplewski, D. C. Hooper, H. Koprowski and V. Yusibov, “Immunization against Rabies with Plant-Derived Antigen,” Proceedings the National Academy of Sciences the USA, Vol. 95, No. 5, 1998, pp. 2481-2485. doi:10.1073/pnas.95.5.2481
- A. H. Cathleen, M. Niezgoda, P. Morrill and C. E. Rupprecht, “Oral Efficacy of an Attenuated Rabies Virus Vaccine in Skunks and Raccoons,” Journal of Wildlife Diseases, Vol. 38, No. 2, 2002, pp. 420-427.
- M. Lee, Y. Zhou, R. Lung, M. L. Chye, W. K. Yip, S. Y. Zee and E. Lam, “Expression of Viral Capsid Protein Antigen against Epstein-Barr Virus in Plastids of Nicotiana tabacum cv. SR1,” Biotechnology and Bioengineering, Vol. 94, No. 6, 2006, pp. 1129-1137. doi:10.1002/bit.20948
- V. Yusibov, A. Modelska, K. Steplewski, M. Agadjanyan, D. Weiner, D. C. Hooper and H. Koprowski, “Antigen Produced in Plants by Infection with Chimeric Plant Viruses Immunize against Rabies Virus and HIV-1,” Proceedings the National Academy of Sciences the USA, Vol. 94, No. 11, 1997, pp. 5784-5788. doi:10.1073/pnas.94.11.5784
- G. G. Zhang, L. Rodrigues, B. Rovinski and K. A. White, “Production of HIV-1 p24 Protein in Transgenic Tobacco Plants,” Molecular Biotechnology, Vol. 20, No. 2, 2002, pp. 131-136. doi:10.1385/MB:20:2:131
- J. Wang and Q. Cui, “Specific Roles of microRNAs in Their Interactions with Environmental Factors,” Journal of Nucleic Acids, Vol. 2012, 2012, Article ID: 978384. doi:10.1155/2012/978384
- F. J. Palella Jr., K. M. Delaney, A. C. Moorman, M. O. Loveless, J. Fuhrer, G. A. Satten, D. J. Aschman and S. D. Holmberg, “HIV Outpatient Study Investigations; Declining Morbidity and Mortality among Patients with Advanced Human Immunodeficiency Virus Infection,” New England Journal of Medicine, Vol. 338, No. 13, 1998, pp. 853-860. doi:10.1056/NEJM199803263381301
- J. Lekakis and I. Ikonomidis, “Cardiovascular Complications of AIDS,” Current Opinion in Critical Care, Vol. 16, No. 5, 2010, pp. 408-412. doi:10.1097/MCC.0b013e32833e10a9
- M. Nunez, “Clinical Syndromes and Consequences of Antiretroviral-Related Hepatotoxicity,” Hepatology, Vol. 52, No. 3, 2010, pp. 1143-1155. doi:10.1002/hep.23716
- C. Xia, D. Luo, X. Yu, S. Jiang and S. Liu, “HIV-Associated Dementia in the Era of Highly Active Antiretroviral Therapy (HAART),” Microbes Infection, Vol. 13, No. 5, 2011, pp. 419-425. doi:10.1016/j.micinf.2011.01.004
- C. Besson, A. Goubar, J. Gabarre, W. Rozenbaum, G. Pialoux, F. P. Châtelet, C. Katlama, F. Charlotte, B. Dupont, N. Brousse, M. Huerre, J. Mikol, P. Camparo, K. Mokhtari, M. Tulliez, D. Salmon-Céron, F. Boué, D. Costaqliola and M. Raphaël, “Changes in AIDS-Related Lymphoma since the Era of Highly Active Antiretroviral Therapy,” Blood, Vol. 98, No. 8, 2001, pp. 2339-2344. doi:10.1182/blood.V98.8.2339
- J. J. Goedert, “The Epidemiology of Acquired Immunodeficiency Syndrome Malignancies,” Seminars in Oncology, Vol. 27, No. 4, 2000, pp. 390-401.
- A. S. Fauci, G. Pantaleo, S. Stanley and D. Weissman, “Immunopathogenic Mechanisms of HIV Infection,” Annals of Internal Medicine, Vol. 124, No. 7, 1996, pp. 654- 663. doi:10.7326/0003-4819-124-7-199604010-00006
- E. Eisele and R. F. Siliciano, “Redefining the Viral Reservoirs that Prevent HIV-1 Eradication,” Immunity, Vol. 37, No. 3, 2012, pp. 377-388.
- C. Kamp, T. Wolf, I. G. Bravo, B. Kraus, B. Krause, B. Neumann, G. Winskowsky, A. Thielen, A. Werner and B. S. Schnierle, “Decreased HIV Diversity after Allogeneic Stem Cell Transplantation of an HIV-1 Infected Patient,” Virology Journal, Vol. 7, 2010, p. 55. doi:10.1186/1743-422X-7-55
- G. Hütter and J. A. Zaia, “Allogeneic Haematopoietic Stem Cell Transplantation in Patients with Human Immunodeficiency Virus: The Experiences of More than 25 Years,” Clinical and Experimental Immunology, Vol. 163, No. 3, 2011, pp. 284-295. doi:10.1111/j.1365-2249.2010.04312.x
- K. Allers, G. Hütter, J. Hofmann, C. Loddenkemper, K. Rieger, E. Thiel and T. Schneider, “Evidence for the Cure of HIV Infection by CCR5∆32/∆32 Stem Cell Transplantation,” Blood, Vol. 117, No. 10, 2011, pp. 2791-2799. doi:10.1182/blood-2010-09-309591
- J. A. Hoxie and C. H. June, “Novel Cell and Gene Therapies for HIV,” Cold Spring Harbor Perspectives in Medicine, Vol. 2, No. 10, 2012, Article ID: a007179.
- Y. R. Fujii, “The Xenotropic microRNA Gene Information for Stem Cell Researches and Clinical Applications,” Stem Cell Discovery, Vol. 3, No. 1, 2013, pp. 32-36. doi:10.4236/scd.2013.31005
- S.-Y. Ying, D. C. Chang and S.-L. Lin, “The microRNA,” Methods in Molecular Biology, Vol. 936, 2013, pp. 1-19. doi:10.1007/978-1-62703-083-0_1
- S. Vasudevan, “Posttranscriptional Upregulation by microRNAs,” Wiley Interdisciplinary Reviews RNA, Vol. 3, No. 3, 2012, pp. 311-330. doi:10.1002/wrna.121
- A. Eulalio, E. Huntzinger and E. Izaurralde, “Getting to the Root of miRNA-Mediated Gene Silencing,” Cell, Vol. 132, No. 1, 2008, pp. 9-14. doi:10.1016/j.cell.2007.12.024
- E. Meiri, A. Levy, H. Benjamin, M. Ben-David, L. Cohen, A. Dov, N. Dromi, E. Elyakim, N. Yerushalmi, O. Zion, G. Lithwick-Yanai and E. Sitbon, “Discovery of microRNAs and Other Small RNAs in Solid Tumors,” Nucleic Acids Research, Vol. 38, No. 18, 2010, pp. 6234-6246. doi:10.1093/nar/gkq376
- S. Djuranovic, M. K. Zinchenko, J. K. Hur, A. Nahvi, J. L. Brunelle, E. J. Rogers and R. Green, “Allosteric Regulation of Argonaute Proteins by miRNAs,” Nature Structural and Molecular Biology, Vol. 17, No. 2, 2010, pp. 144-150.
- S. Djuranovic, A. Nahvi and R. Green, “MiRNA-Mediated Gene Silencing by Translational Repression Followed by mRNA Deadenylation and Decay,” Science, Vol. 336, No. 6078, 2012, pp. 237-240. doi:10.1126/science.1215691
- T. Esposito, S. Magliocca, D. Formicola and F. Gianfrancesco, “PiR_015520 Belongs to Piwi-Associated RNAs Regulates Expression of the Human Melatonin Receptor 1A Gene,” PLoS One, Vol. 6, No. 7, 2011, p. e22727. doi:10.1371/journal.pone.0022727
- F. Zhuang, M. Mastroianni, T. B. White and A. M. Lambowitz, “Linear Group II Intron RNAs Can Retrohome in Eukaryotes and May Use Nonhomologous End-Joining for cDNA Ligation,” Proceedings the National Academy of Sciences the USA, Vol. 106, No. 43, 2009, pp. 18189- 18194. doi:10.1073/pnas.0910277106
- S. A. Krawetz, A. Kruger, C. Lalancette, R. Tagett, E. Anton, S. Draghici and M. P. Diamond, “A Survey of Small RNAs in Human Sperm,” Human Reproduction, Vol. 26, No. 12, 2011, pp. 3401-3412. doi:10.1093/humrep/der329
- L. Chen, J. E. Dahlstrom, S.-H. Lee and D. Rangasamy, “Naturally Occurring Endo-SiRNA Silences LINE-1 Retrotransposons in Human Cells through DNA Methylation,” Epigenetics, Vol. 7, No. 7, 2012, pp. 1-14. doi:10.4161/epi.20706
- T. Mourier and E. Willerslev, “Retrotransposons and NonProtein Coding RNAs,” Brifings in Functional Genomics, Vol. 8, No. 6, 2009, pp. 493-501. doi:10.1093/bfgp/elp036
- J. S. Shapiro, R. A. Langlois, A. M. Pham and B. R. Tenoever, “Evidence for a Cytoplasmic Microprocessor of Pri-miRNAs,” RNA, Vol. 18, No. 7, 2012, pp. 1338- 1346. doi:10.1261/rna.032268.112
- C. F. Althaus, V. Vongrad, B. Niederöst, B. Joos, F. di Giallonardo, P. Rieder, J. Pavlovic, A. Trkola, H. F. Günthard, K. Metzner and M. Fischer, “Tailored Enrichment Strategy Detects Low Abundant Small Noncoding RNAs in HIV-1 Infected Cells,” Retrovirology, Vol. 9, 2012, p. 27. doi:10.1186/1742-4690-9-27
- J. Brennecke, A. Stark, R. B. Russell and S. M. Cohen, “Principles of microRNA-Target Recognition,” PLoS Biology, Vol. 3, 2005, e85.
- A. Grimson, K. K.-H. Farh, W. K. Johnston, L. P. Lim and D. P. Bartel, “MicroRNA Targeting Specificity in Mammals: Determinants beyond Seed Pairing,” Moecular Cell, Vol. 27, No. 1, 2007, pp. 91-105.
- R. C. Friedman, K. K.-H. Farh, C. B. Burge and D. P. Bartel, “Most Mammalian mRNAs Are Conserved Targets of microRNAs,” Genome Research, Vol. 19, No. 1, 2009, pp. 92-105. doi:10.1101/gr.082701.108
- A. A. Bazzini, M. T. Lee and A. J. Giraldez, “Ribosome Profiling Shows that miR-430 Reduces Translation before Causing mRNA Decay in Zebrafish,” Science, Vol. 336, No. 6078, 2012, pp. 233-237. doi:10.1126/science.1215704
- S. Djuranovic, A. Nahvi and R. Green, “A Parsimonious Model for Gene Regulation by miRNAs,” Science, Vol. 331, No. 6017, 2011, pp. 550-553. doi:10.1126/science.1191138
- R. Zhang and B. Su, “MicroRNA Regulation and the Variability of Human Cortical Gene Expression,” Nucleic Acids Research, Vol. 36, No. 14, 2008, pp. 4621-4628.
- N. Li, A. S. Flynt, R. Kim, L. Solnica-Krezel and J. G. Patton, “Dispatched Homolog 2 Is Targeted by miR-214 through a Combination of Three Weak microRNA Recognition Sites,” Nucleic Acids Research, Vol. 36, No. 13, 2008, pp. 4277-4285. doi:10.1093/nar/gkn388
- L. Zhang, D. Hou, X. Chen, D. Li, L. Zhu, Y. Zhang, J. Li, Z. Bian, X. Liang, X. Cai, Y. Yin, C. Wang, T. Zhang, D. Zhu, D. Zhang, J. Xu, J. Zhang, K. Zen and C.-Y. Zhang, “Exogenous Plant MIR168a Specifically Targets Mammalian LDLRAP1: Evidence of Cross-Kingdom Regulation by microRNA,” Cell Research, Vol. 22, No. 1, 2012, pp. 107-126. doi:10.1038/cr.2011.158
- A. Frohn, H. C. Eberl, J. Sthör, E. Glasmacher, S. Rüdel, V. Heissmeyer, M. Mann and G. Meister, “Dicer-Dependent and Independent Argonaute 2 Protein Interaction Network in Mammalian Cells,” Molecular and Cellular Proteomics, Vol. 11, No. 11, 2012, pp. 1442-1456. doi:10.1074/mcp.M112.017756
- Y. R. Fujii, “Formulation of New Algorithmics for miRNAs,” The Open Virology Journal, Vol. 2, 2008, pp. 37-43. doi:10.2174/1874357900802010037
- Y. R. Fujii, “The RNA Gene Information: RetroelementmicroRNA Entangling as the RNA Quantum Code,” Methods in Molecular Biology, Vol. 936, 2013, pp. 47-67. doi:10.1007/978-1-62703-083-0_4
- V. K. Velu, R. Ramesh and A. R. Srinvasan, “Circulating microRNAs as Biomarkers in Health and Disease,” Journal of Clinical and Diagnostic Research, Vol. 6, No. 10, 2012, pp. 1791-1795.
- N. S. Ozek, S. Tuna, A. E. Erson-Bensan and F. Severcan, “Characterization of microRNA-125b Expression in MCF7 Breast Cancer Cells by ATR-FTIR Spectroscopy,” The Analyst, Vol. 135, No. 12, 2010, pp. 3094-3102. doi:10.1039/c0an00543f
- D. Wang, Z. Zhang, E. O'Loughlin, T. Lee, S. Houel, D. O'Carroll, A. Tarakhovsky, N. G. Ahn and R. Yi, “Quantitative Functions of Argonaute Proteins in Mammalian Development,” Genes & Development, Vol. 26, No. 7, 2012, pp. 693-704.
- M. M. Janas, B. Wang, A. S. Harris, M. Aguiar, J. M. Shaffer, Y. V. B. K. Subrahmanyam, M. A. Behike, K. W. Wucherpfennig, S. P. Gygi, E. Gagnon and C. D. Novina, “Alternative RISC Assembly: Binding and Repression of microRNA-mRNA Duplexes by Human Ago Proteins,” RNA, Vol. 18, No. 11, 2012, pp. 2041-2055. doi:10.1261/rna.035675.112
- I. Carmel, N. Shomron and Y. Heifetz, “Does Base-Pairing Strength Play a Role in microRNA Repression?” RNA, Vol. 18, No. 11, 2012, pp. 1947-1956. doi:10.1261/rna.032185.111
- L. Salmena, L. Poliseno, Y. Tay, L. Kats and P. P. Pandolfi, “A ceRNA Hypothesis: The Rosetta Stone of a Hidden RNA Language?” Cell, Vol. 146, No. 3, 2011, pp. 353-358. doi:10.1016/j.cell.2011.07.014
- L. Lipovich, R. Johnson, and C.-Y. Lin, “MacroRNA Underdogs in a microRNA World: Evolutionary, Regulatory, and Biomedical Significance of Mammalian Long Non-Protein-Coding RNA,” Biochemica et Biophysica Acta, Vol. 1799, No. 9, 2010, pp. 597-615. doi:10.1016/j.bbagrm.2010.10.001
- Y. R. Fujii, “Symphony of AIDS: A microRNA-Based Therapy,” In: R. K. Gaur and J. J. Rossi, Eds., Regulation of Gene Expression by Small RNAs, CRC Press, New York, 2009, pp. 333-349. doi:10.1201/9781420008708.ch18
- H. Hartman, “Speculations on the Evolution of the Genetic Code,” Origins of Life, Vol. 6, No. 3, 1975, pp. 423- 427. doi:10.1007/BF01130344
- S. Rodin and S. Ohno, “Four Primordial Modes of tRNASynthetase Recognition, Determined by the (G, C) Operational Code,” Proceedings the National Academy of Sciences the USA, Vol. 94, No. 10, 1997, pp. 5187-5188. doi:10.1073/pnas.94.10.5183
- T. Gu, F. W. Buaas, A. K. Simons, C. L. Ackert-Bicknell, R. E. Braun and M. A. Hibbs, “Canonical A-to-I and C-to-U RNA Editing is Enriched at 3’ UTRs and microRNA Target Sites in Multiple Mouse Tissues,” PLoS One, Vol. 7, 2012, e33720. doi:10.1371/journal.pone.0033720
- A. K. L. Leung, A. G. Young, A. Bhutkar, G. X. Zheng, A. D. Bosson, C. B. Nielsen and P. A. Sharp, “GenomeWide Identification of Ago2 Binding Sites from Mouse Embryonic Stem Cells with and without Mature microRNAs,” Nature Structural and Molecular Biology, Vol. 18, No. 2, 2011, pp. 237-244.
- S. W. Chi, G. J. Hannon and R. B. Darnell, “An Alternative Mode of microRNA Target Recognition,” Nature Structural and Molecular Biolology, Vol. 19, No. 3, 2012, pp. 321-327.
- N. R. Smalheiser and V. I. Torvik, “A Population-Based Statistical Approach Identifies Parameters Characteristic of Human microRNA-mRNA Interactions,” BMC Bioinformatics, Vol. 5, 2004, p. 139. doi:10.1186/1471-2105-5-139
- M. Rossbach, “Non-Coding RNAs in Neural Networks, REST-Assured,” Frontiers in Genetics, Vol. 2, 2011, p. 8. doi:10.3389/fgene.2011.00008
- E. F. Heuston, K. T. Lemon and R. J. Arceci, “The Beginning of the Road for Non-Coding RNAs in Normal Hematopoiesis and Hematologic Malignancies,” Frontiers in Genetics, Vol. 2, 2011, p. 94. doi:10.3389/fgene.2011.00094
- D. Didiano and O. Hobert, “Molecular Architecture of a miRNA-Regulated 3’ UTR,” RNA, Vol. 14, No. 7, 2008, pp. 1297-1317. doi:10.1261/rna.1082708
- P. S. Pang, E. A. Pham, M. Elazar, S. G. Patel, M. R. Eckart and J. S. Glenn, “Structural Map of a microRNA- 122: HCV Complex,” Journal of Virology, Vol. 86, No. 2, 2012, pp. 1250-1254. doi:10.1128/JVI.06367-11
- M. Kertesz, N. Iovino, U. Unnerstall, U. Gaul and E. Segal, “The Role of Site Accessibility in microRNA Target Recognition,” Nature Genetics, Vol. 39, No. 10, 2007, pp. 1278-1284.
- J. Dunningham, K. Burnett, and W. D. Phillips, “BoseEinstein Condensates and Precision Measurements,” Philosophical Transactions. Series A, Mathmatical, Physical, and Engineering Sciences, Vol. 363, No. 1834, 2005, pp. 2165-2175. doi:10.1098/rsta.2005.1636
- J. Estève, C. Gross, A. Weller, S. Giovanazzi and M. K. Oberthaler, “Squeezing and Entanglement in a Bose-Einstein Condensate,” Nature, Vol. 455, No. 7217, 2008, pp. 1216-1219.
- V. Tarallo, Y. Hirano, B. D. Gelfand, B. D. Gelfand, S. Dridi, N. Kerur, Y. Kim, W. G. Cho, H. Kaneko, B. J. Fowler, S. Bogdanovich, R. J. Albuquerque, W. W. Hauswirth, V. A. Chiodo, J. F. Kugel, J. A. Goodrich, S. L. Ponicsan, G. Chaudhuri, M. P. Murphy, J. L. Dunaief, B. K. Ambati, Y. Ogura, J. W. Yoo, D. K. Lee, P. Provost, D. R. Hinton, G. Núńez, M. E. Kleinman and J. Ambati, “DICER 1 Loss and Alu RNA Induce Age-Related Macular Degeneration via the NLRP3 Inflammasome and MyD88,” Cell, Vol. 149, No. 4, 2011, pp. 847-859. doi:10.1016/j.cell.2012.03.036
- J. K. Baillie, M. W. Barnett, K. R. Upton, D. J. Gerhardt, T. A. Richmond, F. de Sapio, P. M. Brennan, P. Rizzu, S. Smith, M. Fell, R. T. Talbot, S. Gustincich, T. C. Freeman, J. S. Mattick, D. A. Hume, P. Heutink, P. Carninci, J. A. Jeddeloh and G. J. Faulkner, “Somatic Retrotransposition Alters the Genetic Landscape of the Human Brain,” Nature, Vol. 479, No. 7374, 2011, pp. 534-537.
- S. Omoto, M. Ito, Y. Tsutsumi, Y. Ichikawa, H. Okuyama, E. A. Brisibe, N. K. Saksena and Y. R. Fujii, “HIV-1 Nef Suppression by Virally Encoded microRNA,” Retrovirology, Vol. 1, 2004, p. 44.
- Y. R. Fujii, “Lost in Translation: Regulation of HIV-1 by microRNAs and a Key Enzyme of RNA-Directed RNA Polymerase,” In: K. Appasani, Ed., MicroRNAs, Cambridge University Press, Cambridge, 2008, pp. 427-442.
- G. Hu, K. M. Drescher and X.-M. Chen, “Exosomal miRNAs: Biological Properties and Therapeutic Potential,” Frontiers in Genetics, Vol. 3, 2012, p. 56. doi:10.3389/fgene.2012.00056
- M. Mittelbrunn and F. Sánchez-Madrid, “Intercellular Communication: Diverse Structures for Exchange of Genetic Information,” Nature Reviews. Molecular Cell Biology, Vol. 13, No. 5, 2012, pp. 328-335.
- M. Chrupek, H. Siipi and L. Martinelli, “Bio-Objects as ‘Boundary Crawlers’: The Case of microRNAs,” Croatian Medical Journal, Vol. 53, No. 3, 2012, pp. 285-288. doi:10.3325/cmj.2012.53.285
- Y. Maida, M. Yasukawa, M. Furuuchi, T. Lassmann, R. Possemato, N. Okamoto, V. Kasim, Y. Hayashizaki, W. C. Hahn and K. Masutomi, “An RNA-Dependent RNA Polymerase Formed by TERT and the RMRP RNA,” Nature, Vol. 461, No. 7261, 2009, pp. 230-235.
- E. A. Gladyshev and I. R. Arkhipova, “A Widespread Class of Reverse Transcriptase-Related Cellular Genes,” Proceedings the National Academy of Sciences the USA, Vol. 108, No. 51, 2011, pp. 20311-20316. doi:10.1073/pnas.1100266108
- D. M. Shechner and D. P. Bartel, “The Structural Basis of RNA-Catalyzed RNA Polymerase,” Nature Structural and Molecular Biology, Vol. 18, No. 9, 2011, pp. 1036- 1042.
- S. Jochum, R. Ruiss, A. Moosmann, W. Hammerschmidt and R. Zeidler, “RNAs in Epstein-Barr Virions Control Early Steps of Infection,” Proceedings the National Academy of Sciences the USA, Vol. 109, No. 21, 2012, pp. E1396-E1404. doi:10.1073/pnas.1115906109
- N. Kosaka, H. Iguchi, Y. Yoshida, K. Hagiwara, F. Takeshita, and T. Ochiya, “Competitive Interactions of Cancer Cells and Normal Cells via Secretory microRNAs,” Journal of Biological Chemistry, Vol. 287, No. 2, 2010, pp. 1397-1405. doi:10.1074/jbc.M111.288662
- Y. Zhang, D. Liu, X. Chen, J. Li, L. Li, Z. Bian, F. Sun, J. Lu, Y. Yin, X. Cai, Q. Sun, K. Wang, Y. Ba, Q. Wang, D. Wang, J. Yang, P. Liu, T. Xu, Q. Yan, J. Zhang and C. Y. Zhang, “Secreted Monocytic miR-150 Enhances Targeted Endothelial Cell Migration,” Molecular Cell, Vol. 39, No. 1, 2010, pp. 133-144. doi:10.1016/j.molcel.2010.06.010
- A. Turchinovich, L. Weiz, A. Langheinz and B. Burwinkel, “Characterization of Extracellular Circulating micro-RNA,” Nucleic Acids Research, Vol. 39, No. 16, 2011, pp. 7223-7233.
- J. D. Arroyo, J. R. Chevillet, E. M. Kroh, I. K. Ruf, C. C. Pritchard, D. F. Gibson, P. S. Mitchell, C. F. Bennett, E. L. Pogosova-Agadjanyan, D. L. Stirewalt, J. F. Tait and M. Tewari, “Argonaute 2 Complex Carry a Population of Circulating microRNAs Independent of Vesicles in Human Plasma,” Proceedings the National Academy of Sciences the USA, Vol. 108, No. 12, 2011, pp. 5003-5008. doi:10.1073/pnas.1019055108
- L. Li, D. Zhu, L. Huang, J. Zhang, Z. Bian, X. Chen, Y. Liu, C.-Y. Zhang and K. Zen, “Argonaute 2 Complexes Selectively Protect the Circulating microRNAs in CellSecreted Microvesicles,” PLoS One, Vol. 7, No. 10, 2012, Article ID: e46957. doi:10.1371/journal.pone.0046957
- Q. Zhou, M. Li, X. Wang, Q. Li, T. Wang, Q. Zhu, X. Zhou, X. Wang, X. Gao and X. Li, ”Immune-Related microRNAs Are Abundant in Breast Milk Exosomes,” International Journal of Biological Sciences, Vol. 8, No. 1, 2012, pp. 118-123. doi:10.7150/ijbs.8.118
- Y. Gu, M. Li, T. Wang, Y. Liang, Z. Zhong, X. Wang, Q. Zhou, L. Chen, Q. Lang, Z. He, X. Chen, J. Gong, X. Gao, X. Li and X. Lv, “Lactation-Related microRNA Expression Profiles of Porcine Breast Milk Exosomes,” PLoS One, Vol. 7, No. 8, 2012, Article ID: e43691. doi:10.1371/journal.pone.0043691
- T. Wurdinger, N. N. Gaston, L. Balaj, B. Kaur, X. O. Breakefielder and D. M. Pegtel, “Extracellular Vesicles and their Convergence with Viral Pathways,” Advances in Virology, Vol. 2012, 2012, Article ID: 767694. doi:10.1155/2012/767694
- T. Fujino and Y. Nagata, “HTLV-I Transmission from Mother to Child,” Journal of Reproductive Immunology, Vol. 47, No. 2, 2000, pp. 197-206.
- S. Martin-Latil, N. Gnädig, A. Mallet, C. Prevost, M. Desdouits, A. Gessain, S. Ozden and P.-E. Ceccaldi, “Mother-to-Child Transmission of HTLV-I: In Vitro Study of HTLV-I Passage across a Tight Human Epithelial Barrier,” Retrovirology, Vol. 6, Suppl. 1, 2009, p. O3. doi:10.1186/1742-4690-6-S1-O3
- K. W. Witwer, A. K. Watson, J. N. Blankson and J. E. Clements, “Relationships of PBMC microRNA Expression, Plasma Viral Load, and CD4+ T-cell Count in HIV- 1-Infected Elite Suppressors and Viremic Patients,” Retrovirology, Vol. 9, 2012, p. 5. doi:10.1186/1742-4690-9-5
- M. Hoque, R. A. Shamanna, D. Guan, T. Pe’ery and M. B. Mathews, “HIV-1 Replication and Latency Are Regulated by Translational Control of Cyclin T1,” Journal of Molecular Biology, Vol. 410, No. 5, 2011, pp. 917-932. doi:10.1016/j.jmb.2011.03.060
- K. Chiang, T.-L. Sung and A. P. Rice, “Regulation of Cyclin T1 and HIV-1 Replication by microRNAs in Resting CD4+ T Lymphocytes,” Journal of Virology, Vol. 86, No. 6, 2012, pp. 3244-3252. doi:10.1128/JVI.05065-11
- C.-J. Shen, Y.-H. Jia, R.-R. Tian, M. Ding, C. Zhang and J.-H. Wang, “Translation of Pur-α Is Targeted by Cellular miRNAs to Modulate the Differentiation-Dependent Susceptibility of Monocytes to HIV-1 Infection,” Official Publication of the Federation of American Societies for Experimental Biology Journal, Vol. 26, No. 11, 2012, pp. 4755-4764. doi:10.1096/fj.12-209023
- L. Houzet, Z. Klase, M. L. Yeung, A. Wu, S.-Y. Le, M. Quiñones and K.-T. Jeang, “The Extent of Sequence Complementarity Correlates with the Potency of Cellular miRNA-Mediated Restriction of HIV-1,” Nucleic Acids Research, Vol. 40, No. 22, 2012, pp. 1168-1196. doi:10.1093/nar/gks912
- N. C. T. Schopman, M. Willemsen, Y. P. Liu, T. Bradley, A. van Kampen, F. Baas, B. Berkhout and J. Haasnoot, “Deep Sequencing of Virus-Infected Cells Reveals HIVEncoded Small RNAs,” Nucleic Acids Research, Vol. 40, No. 1, 2011, pp. 414-427. doi:10.1093/nar/gkr719
- A. Gupta, P. Nagilla, H.-S. Le, C. Bunney, C. Zych, A. Thalamuthu, Z. Bar-Joseph, S. Mathavan and V. Ayyavoo, “Comparative Expression Profile of miRNA and mRNA in Primary Peripheral Blood Mononuclear Cells Infected with Human Immunodeficiency Virus (HIV-1),” PLoS One, Vol. 6, No. 7, 2011, Article ID: e22730. doi:10.1371/journal.pone.0022730
- Z. Klase, L. Houzet and K.-T. Jeang, “Replication Competent HIV-1 Viruses that Express Intragenomic microRNA Reveal Discrete RNA-Interference Mechanisms that Affect Viral Replication,” Cell and Bioscience, Vol. 1, No. 1, 2011, p. 38. doi:10.1186/2045-3701-1-38
- N. H. Gana, T. Onuki, A. F. B. Victoriano and T. Okamoto, “MicroRNAs in HIV-1 Infection: An Integration of Viral and Cellular Interaction at the Genomic Level,” Frontiers in Microbiology, Vol. 3, 2012, p. 306. doi:10.3389/fmicb.2012.00306
- K. Chiang, H. Liu and A. P. Rice, “MiR-132 Enhaces HIV-1 Replication,” Virology, Vol. 438, No. 1, 2013, pp. 1-4.
- V. Soriano, P. Barreiro, L. Martin-Carbonero, C. Castellares, A. Ruiz-Sancho, P. Labarga, B. Ramos and J. Gonzalez-Lahoz, “Treatment of Chronic Hepatitis B or C in HIV-Infected Patients with Dual Viral Hepatitis,” The Journal of Infectious Diseases, Vol. 195, No. 8, 2007, pp. 1181-1183. doi:10.1086/512679
- C. Esau, S. Davis, S. F. Murray, X. X. Yu, S. K. Pandey, M. Pear, L. Watts, S. L. Booten, M. Graham, R. McKay, A. Subramaniam, S. Propp, B. A. Lollo, S. Freier, C. F. Bennett, S. Bhanot and B. P. Monia, “MiR- Regulation of Lipid Metabolism Revealed by in Vivo Antisense Targeting,” Cell Metabolism, Vol. 3, No. 2, 2006, pp. 87- 98.
- D. Gatfield, G. Le Martelot, C. E. Vejnar, D. Gerlach, O. Schaad, F. Fleury-Olela, A.-L. Ruskeepää, M. Oresic, C. C. Esau, E. M. Zdobnov and U. Schibler, “Intergration of microRNA miR-122 in Hepatic Circadian Gene Expression,” Genes and Development, Vol. 23, No. 11, 2012, pp. 1313-1326. doi:10.1101/gad.1781009
- S.-H. Hsu, B. Wang, J. Kota, J. Yu, S. Costinean, H. Kutay, L. Yu, S. Bai, K. La Perle, R. R. Chivukula, H. Mao, M. Wei, K. R. Clark, J. R. Mendell, M. A. Caliguri, S. T. Jacob, J. T. Mendell and K. Ghoshal, “Essential Metabolic, Anti-Inflammatory, and Anti-Tumorigenic Functions of miR-122 in Liver,” The Journal of Clinical Investigation, Vol. 122, No. 8, 2012, pp. 2871-2883. doi:10.1172/JCI63539
- C. L. Jopling, M. Yi, A. M. Lancaster, S. M. Lemon and P. Sarnow, “Modulation of Hepatitis C Virus RNA Abundance by a Liver-Specific microRNA,” Science, Vol. 309, No. 5740, 2005, pp. 1577-1581. doi:10.1126/science.1113329
- C. L. Jopling, “Regulation of Hepatitis C Virus by microRNA-122,” Biochemical Society Transactions, Vol. 36, No. 6, 2008, pp. 1220-1223. doi:10.1042/BST0361220
- J. I. Henke, D. Goergen, J. Zheng, Y. Song, C. G. Schüttler, C. Fehr, C. Jünemann and M. Niepmann, “MicroRNA-122 Stimulates Translation of Hepatitis C Virus RNA,” The EMBO Journal, Vol. 27, No. 24, 2008, pp. 3300-3310.
- K. L. Norman and P. Sarnow, “Modulation of Hepatitis C Virus RNA Abundance and the Isoprenoid Biosynthesis Pathway by microRNA miR-122 Involves Distinct Mechanisms,” Journal of Virology, Vol. 84, No. 1, 2010, pp. 666-670. doi:10.1128/JVI.01156-09
- R. A. Villanueva, R. K. Jangra, M. Yi, R. Pyles, N. Bourne and S. M. Lemon, “MiR-122 Does Not Modulate the Elongation Phase of Hepatitis C Virus RNA Synthesis in Isolated Replicase Complexes,” Antiviral Research, Vol. 88, No. 1, 2010, pp. 119-123. doi:10.1016/j.antiviral.2010.07.004
- M. Lindow and S. Kauppinen, “Discovering the First microRNA-Target Drug,” The Journal of Cell Biology, Vol. 199, No. 3, 2012, pp. 407-412. doi:10.1083/jcb.201208082
- M. Sarasin-Filipowicz, J. Krol, I. Markiewicz, M. H. Heim and W. Filipowicz, “Decreased Levels of microRNA miR-122 in Individuals with Hepatitis C Responding Poorly to Interferon Therapy,” Nature Medicine, Vol. 15, No. 1, 2009, pp. 31-33.
- S. Wang, L. Qiu, X. Yan, W. Jin, Y. Wang, L. Chen, E. Wu, X. Ye, G. F. Gao, F. Wang, Y. Chen, Z. Duan and S. Meng, “Loss of microRNA 122 Expression in Patients with Hepatitis B Enhances Hepatitis B Virus Replication through Cyclin G1-Modulated P53 Activity,” Hepatology, Vol. 55, No. 3, 2012, pp. 730-741. doi:10.1002/hep.24809
- Z. Y. Li, Y. Xi, W. N. Zhu, C. Zeng, Z. Q. Zhang, Z. C. Guo, D. L. Hao, G. Liu, L. Feng, H. Z. Chen, F. Chen, X. Lv, D. P. Liu and C. C. Liang, “Positive Regulation of Hepatic miR-122 Expression by HNF-4 Alpha,” Journal of Hepatology, Vol. 55, No. 3, 2011, pp. 602-611. doi:10.1016/j.jhep.2010.12.023
- R. Triboulet, B. Mari, Y. L. Lin, C. Chable-Bessia, Y. Bennasser, K. Lebrigand, B. Cardinaud, T. Maurin, P. Barbry, V. Baillat, J. Reynes, P. Corbeau, K. T. Jeang and M. Benkirane, “Suppression of microRNA-Silencing Pathway by HIV-1 during Virus Replication,” Science, Vol. 315, No. 5818, 2007, pp. 1579-1582. doi:10.1126/science.1136319
- A. Khokhar, S. Noorali, M. Sheraz, K. Mahalingham, D. G. Pace, M. R. Khanani and O. Bagasra, “Computational Analysis to Predict Functional Role of hsa-miR-3065-3p as an Antiviral Therapeutic Agent for Treatment of Triple Infections: HCV, HIV-1, and HBV,” The Libyan Journal of Medicine, Vol. 7, 2012, p. 19774. doi:10.3402/ljm.v7i0.19774
- P. Garred, “Chemokine-Receptor Polymorphisms: Clarity or Confusion for HIV-1 Prognosis?” The Lancet, Vol. 351, No. 9095, 1998, pp. 2-3.
- Y. Huang, W. A. Paxton, S. M. Wolinsky, A. U. Neumann, L. Zhang, T. He, S. Kang, D. Ceradini, Z. Jin, K. Yazdanbakhsh, K. Kunstman, D. Erickson, E. Dragon, N. R. Landau, J. Phair, D. D. Ho and R. A. Koup, “The Role of a Mutant CCR5 Allele in HIV-1 Transmission and Disease Progression,” Nature Medicine, Vol. 2, No. 11, 1996, pp. 1240-1243. doi:10.1038/nm1196-1240
- P. Jungebluth, E. Alici, S. Baiguera, K. Le Blanc, P. Blomberg, B. Bozóky, C. Crowley, O. Einarsson, K. H. Grinnemo, T. Gudbjartsson, S. Le Guyader, G. Henriksson, O. Hermanson, J. E. Juto, B. Leidner, T. Lilja, J. Liska, T. Luedde, V. Lundin, G. Moll, B. Nilsson, C. Roderburg, S. Strõmblad, T. Sutlu, A. I. Teixeira, E. Watz, A. Seifalian and P. Macchiarini, “Tracheobronchial Transplantation with a Stem-Cell-Seeded Bioartificial Nanocomposite: A Proof-of-Concept Study,” The Lancet, Vol. 378, No. 9808, 2011, pp. 1997-2004. doi:10.1016/S0140-6736(11)61715-7
- D. Ganten, “What is life? On Erwin Schrödinger, His Cat, and the Journal of Molecular Medicine,” Journal of Molecular Medicine, Vol. 85, No. 12, 2007, pp. 1291-1292. doi:10.1007/s00109-007-0288-9
- J. T. Trevors and L. Masson, “Quantum Microbiology,” Current Issues in Molecular Biology, Vol. 13, No. 2, 2011, pp. 43-49.
- G. P. Berman, G. D. Doolen, R. Mainieri and V. I. Tsifrinovich, “Introduction to Quantum Computers,” World Scientific Pub. Co. Ltd., Singapore City, 1998.
- T. Yamamoto, S. Omoto, M. Mizuguchi, H. Mizukami, H. Okuyama, N. Okada, N. K. Saksena, E. A. Brisibe, K. Otake and Y. R. Fujii, “Double-Stranded Nef RNA Interferes with Human Immunodeficiency Virus Type 1 Replication,” Microbiology and Immunology, Vol. 46, No. 11, 2002, pp. 809-817.

