Open Journal of Orthopedics
Vol. 2 No. 2 (2012) , Article ID: 20118 , 7 pages DOI:10.4236/ojo.2012.22013
Use of Synteny Conversion in Identification of Candidate Genes for Somitogenesis in Humans
![]()
1University of Wisconsin, Madison, USA; 2Hospital for Special Surgery, New York, USA.
Email: pfgiampietro@pediatrics.wisc.edu
Received March 2nd, 2012; revised April 5th, 2012; accepted May 13th, 2012
Keywords: Synteny; Candidate Genes; Vertebral Patterning Genes; Congenital Scoliosis; Idiopathic Scoliosis
ABSTRACT
Understanding the genetic component of scoliosis in humans has relied on the assumption that spine development is conserved across species. Since evolutionary conserved genes tend to lie within synteny blocks (HSBs) and genes which are not conserved lie within evolutionary breakpoint regions (EBRs), HSB analysis may be used to determine if spine development is conserved across species. We hypothesized that vertebral patterning genes are conserved in amniotes and their location is within stable or “syntenic” regions of chromosomes. Seventy seven patterning genes involved in Fgf, Wnt and Notch signaling pathways were analyzed to determine their location within HSBs or EBRs in the genomes of several amniotic species. The human genome was divided into 1 Mbp intervals and a comparison was made to determine whether these genes were preferentially localized within HSBs or EBRs associated with rapid evolution. The results indicate that genes associated with somite development in humans are preferentially located away from the EBRs: 0.014 genes in EBRs on genome average vs. 0.030 on average in other parts of the genome (p-value = 0.01). The concentration of vertebral patterning genes in HSBs, provides evidence that developmental pathways involved in vertebral morphogenesis are likely conserved across amniotes, consistent with their known function. These data support prior observations indicating that gene networks associated with major developmental processes such as neuronal, central nervous system, bone and blood vessel development, some mediated by Wnt and Notch signaling pathways, were less likely to be localized at EBRs.
1. Introduction
Congenital and idiopathic scoliosis constitute two major categories of spinal curvature. Idiopathic scoliosis, as defined by the Scoliosis Research Society refers to a lateral curvature of 10˚ or greater on plane radiographs, is not associated with any underlying cause [1]. Congenital scoliosis is a spinal curvature that results from developmental abnormalities in vertebral bodies, which are referred to as congenital vertebral malformations (CVM). These abnormalities may further be divided into disorders of formation (wedge vertebrae, hemivertebrae) or disorders of segmentation (vertebral bar). Both congenital and idiopathic scoliosis are clinically and etiologically heterogeneous. Although the genetic mechanisms responsible for both conditions are not well understood, there is an observed prevalence of 17.3% of congenital scoliosis in families with idiopathic scoliosis, suggesting similar underlying pathogenic mechanisms [2].
Mutations in genes associated with somitogenesis represent ideal candidates for scoliosis. Mesodermally derived somites are paired structures that give rise to the vertebral bodies, ribs, spinal and rib associated muscles and tendons. Somitogenesis occurs by an intricate interplay of patterning genes which are expressed in the presomitic mesoderm. Much of our understanding of the somitogenesis process has been derived from experimental work in mice, chicken and Xenopus species. The Notch, Fgf and Wnt signaling pathways regulated by a molecular oscillator in which the Notch and Fgf genes oscillate in opposite phase to the Wnt genes [3]. Since a large number of human disorders are characterized by aberrant spine development including congenital scoliosis, spondylocostal dysostosis, spondylothoracic dysostosis, Klippel-Feil syndrome (fusion of cervical vertebrae, short neck), hemifacial microsomia (ear tags, microtia, cardiac abnormalities, vertebral abnormalities) and VACTERL syndrome (vertebral anomalies, anal atresia, cardiac abnormalities, tracheo-esophageal fistula, renal abnormalities, limb abnormalities) the clinical relevance for understanding the genetic basis of somitogenesis becomes important. Several prior studies have incorporated a candidate gene approach based on the assumption that human genes that are syntenic to mouse genes are associated with spine development [3-9].
There are inherent difficulties in the identification of genes contributing to scoliosis in humans. Most cases of congenital vertebral malformations (CVMs) represent sporadic occurrences within a single family, thus making traditional linkage approaches difficult to utilize. The large number of potential candidate genes to choose from, compounded by a clinical heterogeneity of CVM phenoltypes, makes this a difficult area to provide genetic diagnosis and counseling for families.
Multiple factors may contribute to the development of idiopathic scoliosis including muscle imbalance and changes in the connective tissue matrix. Linkage and association studies have identified a number of genetic regions associated with idiopathic scoliosis [10-13]. Polymorphisms in CHD7, a chromeodomain helicase which is associated with CHARGE syndrome (Coloboma, Atresia Choanae, Retarded Growth, Ear Anomalies), have been associated with idiopathic scoliosis [11]. CHD7 is associated with embryonic axial development in mice, providing additional evidence that congenital and idiopathic scoliosis may have a unifying pathogenetic mechanism [14].
Prior studies have demonstrated that considerable evolutionary activity exists at the evolutionary breakpoint regions (EBRs) which are located between homologous synteny blocks (HSBs) including reuse, increased gene density, segmental duplication accumulation and the emergence of centromeres and telomeres [15]. EBR is defined as an interval between two adjacent HSBs that is demarcated by the end-sequence coordinates of those HSBs on each side. Because the process of spinal column development is similar among amniotes, we hypothesized that genes associated with scoliosis are conserved in amniotes and their location is within the regions of conserved synteny of chromosomes in different mammals.
2. Methods
Ninety seven patterning genes including genes from the Wnt, Fgf, and Notch signaling pathways in addition to other patterning genes operative in mice somitogenesis and associated with scoliosis phenotypes, were initially identified for synteny block analyses [3]. The analysis was performed in order to determine whether these genes involved in somitogenesis and scoliosis are preferentially located in the regions of mammalian chromosomes that are stable in evolution, or whether they are located in the regions that correspond to positions of EBRs in the genomes of several amniotic species (human, chimp, macaque, mouse, rat, dog, pig, cattle, opossum, chicken).
Mouse gene coordinates corresponding to the human chromosome coordinates for the 77 genes from Table 1 (20 of the original 97 patterning genes did not have corresponding mouse coordinates) were obtained by using Ensemble homology tables [16]. The human genome was divided into 2980, 1 Mbp intervals; the number of the genes from Table 1 was counted in each of those intervals. A determination was made as to which bins are overlapping with positions of the HSBs or EBRs. Student’s t-test analysis with unequal variances, as described previously, was performed to determine whether the somite patterning genes are preferentially located in the EBRs or HSBs [17,18].
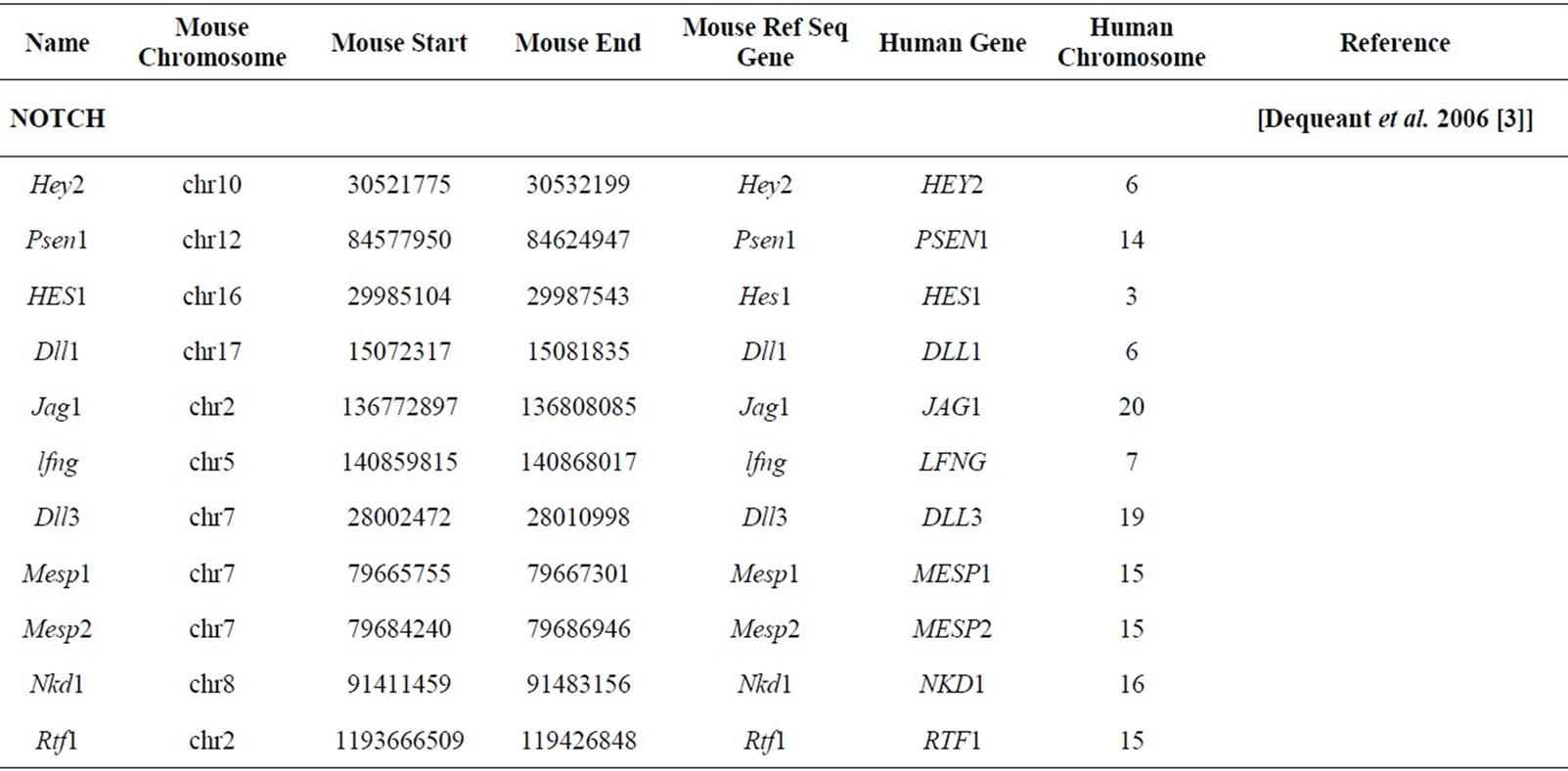



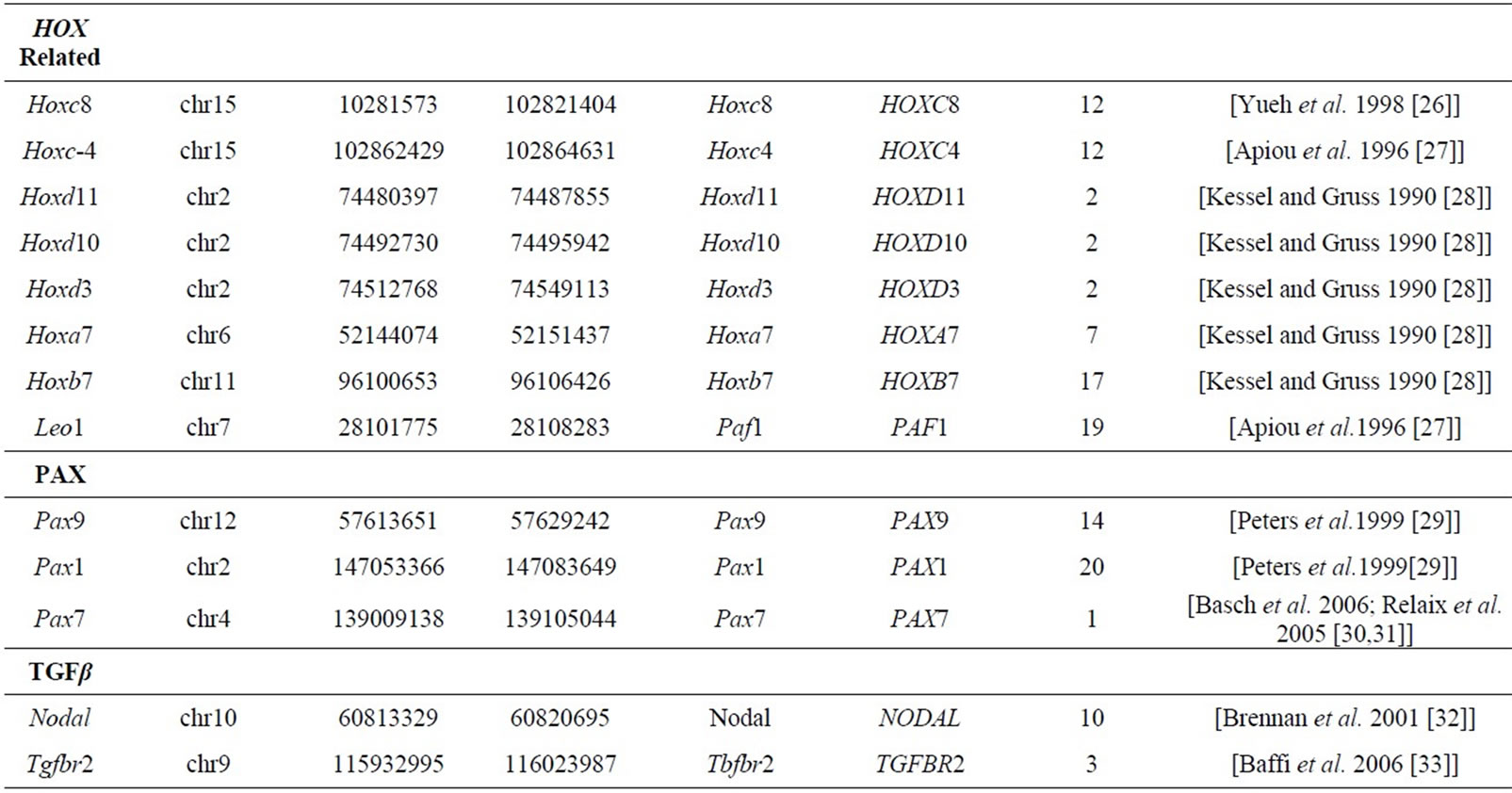

Table 1. Mouse genes and coordinates studied with corresponding human syntenic gene region.
3. Results
Vertebral patterning and scoliosis associated genes in Table 1 were found to be preferentially located away from the EBRs, with approximately twice as many genes on average occurring in other parts of the genome as compared to the breakpoint intervals (p-value = 0.011). While this does not appear to be a large difference, if all genes in the genome are counted, the EBRs on average contain ~2 times more genes than the rest of the genome. In general, breakpoint intervals are significantly enriched for genes, and the results of this analysis indicate that they are not enriched for vertebral patterning genes. Examination of large blocks (>3 Mb) of homologous synteny (approximately 7 of these occur in amniote genomes, which are
>16.3 Mbp in human coordinates) indicated 0.04 genes from Table 1 localized in these blocks on genome average, while 0.03 genes localized to the rest of the genome [19]. This result is not statistically significant, probably because of the small number of genes in this comparison.
4. Discussion
Synteny block analysis performed on 77 genes associated with Wnt, Fgf and Notch signaling pathways indicated that these genes are located away from the boundaries of EBRs. The location of vertebral patterning genes away from synteny breakpoints highlights their important and conserved evolutionary function in amniotes. This is the first study to analyze conserved synteny for genes associated with somitogenesis in amniotic species and provides additional genetic evidence for similarities in spine development in amniotes.
A prior analysis of mouse scoliotic phenotypes using the Mouse Genome Database (MGD), followed by use of the Online Mendelian Inheritance in Man (OMIM), yielded 45 genes with possible scoliosis phenotypes. Twenty eight genes were translated to the human genome coordinates using mouse and human synteny maps [8]. These included WNT3A and DLL3 genes, also members of the cohort of genes in Table 1. During this analysis it was not possible to determine whether each vertebral patterning gene was located within EBRs or away from breakpoint intervals. The localization of patterning genes associated with human vertebral development to regions away from synteny breakpoint intervals provides evidence for conservation of the basic vertebral patterning scheme during amniote development.
These data are consistent with prior analyses performed by Larkin et al. [19]. Gene networks associated with major developmental processes such as neuronal development, central nervous system, bone and blood vessel development, some of which were mediated by Wnt and Notch signaling pathways, were significantly enriched in HSBs and, therefore, less likely to be localized at EBRs. Gene networks associated with responses to external stimuli such as inflammatory responses and muscle contraction were more likely to be localized to EBRs. Our study focused on 77 genes associated with somitogenesis including 11 NOTCH, 12 FGF, 29 WNT, 8 HOX, 3 PAX, 2 TGFβ pathway and 12 additional genes associated with scoliosis phenotypes and the results demonstrated these patterning genes were significantly overrepresented in the evolutionary conserved regions.
Using a series of bioinformatic approaches including neighbor-joining (NJ) and maximum parsimony (MP), contained within the PHYLIP (PHYLogeny inference package) software package [20], the evolution of Notch family proteins in species from worm to human was analyzed in C. elegans, D. melanogaster, C. intestinalis, and H. sapiens using Mapviewer, Geneview, and the BlastP and TBlastN algorithm [21]. The chromosomal distribution of PBX (pre-B cell leukemia homeobox), LHX3 (Lim homeobox 3), NRARP (Notch regulated ankyrin repeat protein), BRD (bromodomain) and CAMSAP1 (calmodulin regulated spectrin-associated protein 1) was found to follow the distribution of Notch, providing evidence for co-evolution with Notch signaling pathway genes by segmental duplication. The close proximity of these genes may reflect a functional relationship. For instance, in C. elegans, PBX appears to be responsible for transcriptional control of Notch signaling [22].
This study has several limitations. While genes in the FGF, WNT and Notch signaling pathways were analyzed, genes in other pathways such as the BMP signaling pathway were not studied. Corresponding mouse coordinates were identified for 77 of the 97 patterning human genes originally identified. Due to the relatively small number of genes studied, it was not possible to determine whether the vertebral patterning genes were preferentially localized to large blocks of homologous synteny.
Besides playing a crucial role in somitogenesis, the Fgf, Wnt and Notch signaling pathways also are involved in the embryogenesis of other organs. Fgf’s have important roles in development of the limbs, skin, central nervous system, ear, lungs, liver and have major involvement in the wound healing process [23]. Fibroblast growth factor receptor related disorders in humans include craniosynostosis syndromes such as Apert and Crouzon syndrome, and skeletal dsyplasias, of which achondroplasia is the most common. The Wnt canonical pathway is active in neural tube development [24]. Notch signaling is involved in developmental pathways which affect the vasculature, heart, eye and liver [25]. Both Notch and Wnt pathways are involved in autonomous phenotypes including cellular development and proliferation. It is possible that the conservation of Fgf, Wnt and Notch signaling related genes in HSBs reflects conservation of non-somite signaling functions or conservation of multiple signaling functions. A previous study by Larkin et al. focused on association of HSBs with respect to gene networks, while our study was aimed at localization of genes associated with a specific disease process, namely scoliosis with respect to HSBs [19].
In summary we provide further evidence that developmental pathways associated with somitogenesis are conserved across amniotes which is consistent with their known function.
5. Acknowledgements
We appreciate the comments and advice provided by Drs. Harris Lewin and Denis Larkin.
REFERENCES
- Scoliosis Research Society, “Glossary of Scoliosis Terms,” Spine, Vol. 1, No. 1, 1976, pp. 57-58. doi:10.1097/00007632-197603000-00008
- S. B. Purkiss, B. Driscoll, W. G. Cole and B. Alman, “Idiopathic Scoliosis in Families of Children with Congenital Scoliosis,” Clinical Orthopaedics and Related Research, Vol. 401, 2002, pp. 27-31. doi:10.1097/00003086-200208000-00005
- M. Dequeant, E. Glynn, K. Gaundenz, M. Wahl, J. Chen, A. Mushegian and O. Pourquie, “A Complex Oscillating Network of Signaling Genes Underlies the Mouse Segmentation Clock,” Science, Vol. 314, No. 5805, 2006, pp. 1595-1598. doi:10.1126/science.1133141
- N. Ghebranious, R. Blank, C. Raggio, J. Staubli, E. McPherson, L. Ivacic, K. Rasmussen, F. Jacobsen, T. Faciszewski, J. K. Burmester, R. M. Pauli, O. BoachieAdjei, I. Glurich and P. F. Giampietro, “A Missense T (Brachyury) Mutation Contributes to Vertebral Malformations,” Journal of Bone and Mineral Research, Vol. 23, No. 10 2008, pp. 1576-1583. doi:10.1359/jbmr.080503
- N. Ghebranious, J. Burmester, I. Glurich, E. McPherson, L. Ivacic, J. Kislow, K. Rasmussen, V. Kumar, C. Raggio, R. Blank, F. S. Jacobsen, T. Faciszewski, J. Womack and P. F. Giampietro, “Evaluation of SLC35A3 as a Candidate Gene for Human Vertebral Malformations,” American Journal of Medical Genetics, Vol. 140A, No. 12, 2006, pp. 1346-1348. doi:10.1002/ajmg.a.31307
- N. Ghebranious, C. L. Raggio, R. D. Blank, E. McPherson, J. K. Burmester, L. Ivacic, K. Rasmussen, J. Kislow, I. Glurich, F. S. Jacobsen, T. Faciszewski, R. M. Pauli, O. Boachie-Adjei and P. F. Giampietro, “Lack of Evidence of WNT3A as a Candidate Gene for Congenital Vertebral Malformations,” Scoliosis, Vol. 2, 2007, p. 13. doi:10.1186/1748-7161-2-13
- P. Giampietro, C. Raggio, C. Reynolds, N. Ghebranious, J. Burmester, I. Glurich, K. Rasmussen, E. McPherson, R. Pauli, S. K. Shukla, S. Merchant, F. S. Jacobsen, T. Faciszewski and R. D. Bland, “DLL3 as a Candidate for Vertebral Malformations,” American Journal of Medical Genetics, Vol. 140A, No. 22, 2006, pp. 2447-2453. doi:10.1002/ajmg.a.31509
- P. F. Giampietro, C. L. Raggio and R. D. Blank, “SyntenyDefined Candidate Genes for Idiopathic and Congenital Scoliosis,” American Journal of Medical Genetics, Vol. 83, No. 3, 1999, pp. 164-177. doi:10.1002/(SICI)1096-8628(19990319)83:3<164::AID-AJMG5>3.0.CO;2-D
- P. F. Giampietro, C. L. Raggio, C. E. Reynolds, S. K. Shukla, E. McPherson, N. Ghebranious, F. S. Jacobsen, V. Kumar, T. Faciszewski, R. M. Pauli, K. Rasmussen, J. K. Burmester, C. Zaleski, S. Merchant, D. David, J. L. Weber, I. Glurich and R. D. Blank, “An Analysis of PAX1 in the Development of Vertebral Malformations,” Clinical Genetics, Vol. 68, No. 5, 2005, pp. 448-453. doi:10.1111/j.1399-0004.2005.00520.x
- K. J. Alden, B. Marosy, N. Nzegwu, C. M. Justice, A. F. Wilson and N. H.Miller, “Idiopathic Scoliosis: Identification of Candidate Regions on Chromosome 19, p13,” Spine, Vol. 31, No. 16, 2006, pp. 1815-1819. doi:10.1097/01.brs.0000227264.23603.dc
- X. Gao, D. Gordon, D. Zhang, R. Browne, C. Helms, J. Gillum, S. Weber, S. Devroy, S. Swaney, M. Dobbs, J. Morcuende, V. Sheffield, M. Lovett, A. Bowcock, J. Herring and C. Wise, “CHD7 Gene Polymorphisms Are Associated with Susceptibility to Idiopathic Scoliosis,” American Journal of Human Genetics, Vol. 80, No. 5, 2007, pp. 957-965. doi:10.1086/513571
- N. H. Miller, C. M. Justice, B. Marosy, K. F. Doheny, E. Pugh, J. Zhang, H. C. Dietz 3rd and A. F. Wilson, “Identification of Candidate Regions for Familial Idiopathic Scoliosis,” Spine, Vol. 30, No. 10, 2005, pp. 1181-1187. doi:10.1097/01.brs.0000162282.46160.0a
- N. H. Miller, B. Marosy, C. M. Justice, S. M. Novak, E. Y. Tang, P. Boyce, J. Pettengil, K. F. Doheny, E. W. Pugh and A. F. Wilson, “Linkage Analysis of Genetic Loci for Kyphoscoliosis on Chromosomes 5, p13,” 13q13.3, and 13q32,” American Journal of Medical Genetics A, Vol. 140, No. 10, 2006, pp. 1059-1068. doi:10.1002/ajmg.a.31211
- V. Subramanian, B. I. Meyer and P. Gruss, “Disruption of the Murine Homeobox Gene Cdx1 Affects Axial Skeletal Identities by Altering the Mesodermal Expression Domains of Hox Genes,” Cell, Vol. 83, No. 4, 1995, pp. 641-653. doi:10.1016/0092-8674(95)90104-3
- W. J. Murphy, D. M. Larkin, A. E. van der Wind, G. Bourque, G. Tesler, L. Auvil, J. E. Beever, B. P. Chowdhary, F. Galibert, L. Gatzke, C. Hitte, S. N. Meyers, D. Milan, E. A. Ostrander, G. Pape, H. G. Parker, T. Raudsepp, M. B. Roqatcheva, L. B. Schook, L. C. Skow, M. Welge, J. E. Womack, S. J. O’brien, P. A. Pevzner and H. A. Lewin, “Dynamics of Mammalian Chromosome Evolution Inferred from Multispecies Comparative Maps,” Science, Vol. 309, No. 5734, 2005, pp. 613-617. doi:10.1126/science.1111387
- ensemble.org
- E. Skovlund and G. U. Fenstad, “Should We Always Choose a Nonparametric Test When Comparing Two Apparently Nonnormal Distributions?” Journal of Clinical Epidemiology, Vol. 54, No. 1, 2001, pp. 86-92. doi:10.1016/S0895-4356(00)00264-X
- A. V. Smith, D. J. Thomas, H. M. Munro and G. R. Abecasis, “Sequence Features in Regions of Weak and Strong Linkage Disequilibrium,” Genome Research, Vol. 15, No. 11, 2005, pp. 1519-1534. doi:10.1101/gr.4421405
- D. M. Larkin, G. Pape, R. Donthu, L. Auvil, M. Welge and H. A. Lewin, “Breakpoint Regions and Homologous Synteny Blocks in Chromosomes Have Different Evolutionary Histories,” Genome Research, Vol. 19, No. 5, 2009, pp. 770-777. doi:10.1101/gr.086546.108
- evolution.gs.org
- A. Theodosiou, S. Arhondakis, M. Baumann and S. Kossida, “Evolutionary Scenarios of Notch Proteins,” Molecular Biology and Evolution, Vol. 26, No. 7, 2009, pp. 1631-1640. doi:10.1093/molbev/msp075
- K. Takacs-Vellai, T. Vellai, E. B. Chen, Y. Zhang, F. Guerry, M. J. Stern and F. Muller, “Transcriptional Control of Notch Signaling by a HOX and a PBX/EXD Protein during Vulval Development in C. elegans,” Developmental Biology, Vol. 302, No. 2, 2007, pp. 661-669. doi:10.1016/j.ydbio.2006.09.049
- X. Coumoul and C. X. Deng, “Roles of FGF Receptors in Mammalian Development and Congenital Diseases,” Birth Defects Research C: Embryo Today, Vol. 69, No. 4, 2003, pp. 286-304. doi:10.1002/bdrc.10025
- R. Alvarez-Medina, J. Cayuso, T. Okubo, S. Takada and E. Marti, “Wnt Canonical Pathway Restricts Graded Shh/Gli Patterning Activity through the Regulation of Gli 3 Expression,” Development, Vol. 135, 2008, pp. 237- 247. doi:10.1242/dev.012054
- T. Gridley, “Notch Signaling and Inherited Human Disease Syndromes,” Human Molecular Genetics, Vol. 12, No. 1, 2003, pp. R9-R13. doi:10.1093/hmg/ddg052
- Y. G. Yueh, D. P. Gardner and C. Kappen, “Evidence for Regulation of Cartilage Differentiation by the Homeobox Gene Hoxc-8,” Proceedings of the National Academy of Sciences of the United States of America, Vol. 95, No. 17, 1998, pp. 9956-9961. doi:10.1073/pnas.95.17.9956
- F. Apiou, D. Flagiello, C. Cillo, B. Malfoy, M. F. Poupon and B. Dutrillaux, “Fine Mapping of Human HOX Gene Clusters,” Cytogenetics and Cell Genetics, Vol. 73, No. 1-2, 1996, pp. 114-115. doi:10.1159/000134320
- M. Kessel and P. Gruss, “Murine Developmental Control Genes,” Science, Vol. 249, No. 4967, 1990, pp. 374-379. doi:10.1126/science.1974085
- H. Peters, B. Wilm, N. Sakai, K. Imai, R. Mass and R. Balling, “Pax1 and Pax9 Synergistically Regulate Vertebral Column Development,” Development, Vol. 126, 1999, pp. 5399-5408.
- M. L. Basch, M. Bronner-Fraser and M. I. Garcia-Castro, “Specification of the Neural Crest Occurs during Gastrulation and Requires Pax7,” Nature, Vol. 441, No. 7090, 2006, pp. 218-222. doi:10.1038/nature04684
- F. Relaix, D. Rocancourt, A. Mansouri and M. Buckingham, “A Pax3/Pax7-Dependent Population of Skeletal Muscle Progenitor Cells,” Nature, Vol. 435, No. 7044, 2005, pp. 948-953. doi:10.1038/nature03594
- J. Brennan, C. C. Lu, D P. Norris, T. A. Rodriguez, R. S. Beddington and E. J. Robertson, “Nodal Signaling in the Epiblast Patterns the Early Mouse Embryo,” Nature, Vol. 411, No. 6840, 2001, pp. 965-969. doi:10.1038/35082103
- M. O. Baffi, M. A. Moran and R. Serra, “Tgfbr2 Regulates the Maintenance of Boundaries in the Axial Skeleton,” Developmental Biology, Vol. 296, No. 2, 2006, pp. 363-374. doi:10.1016/j.ydbio.2006.06.002
- J. T. Eggenschwiler, E. Espinoza and K. V. Anderson, “Rab23 Is an Essential Negative Regulator of the Mouse Sonic Hedgehog Signalling Pathway,” Nature, Vol. 412, No. 6843, 2001, pp. 194-198. doi:10.1038/35084089
- A. Vortkamp, K. Lee, B. Lanske, G. V. Segre, H. M. Kronenberg and C. J. Tabin, “Regulation of Rate of Cartilage Differentiation by Indian Hedgehog and PTH-Related Protein [See Comments],” Science, Vol. 273, No. 5275, 1996, pp. 613-622. doi:10.1126/science.273.5275.613
- T. Kanda, Y. Yoshida, Y. Izu, A. Nifuji, Y. Ezura, K. Nakashima and M. Noda, “PlexinD1 Deficiency Induces Defects in Axial Skeletal Morphogenesis,” Journal of Cellular Biochemistry, Vol. 101, No. 6, 2007, pp. 1329- 1337. doi:10.1002/jcb.21306
- P. Bialek, B. Kern, X. Yang, M. Schrock, D. Sosic, N. Hong, H. Wu, K. Yu, D. M. Ornitz, E. N. Olson, M. J. Justice and G. Karsenty, “A Twist Code Determines the Onset of Osteoblast Differentiation,” Developmental Cell, Vol. 6, No. 3, 2004, pp. 423-435. doi:10.1016/S1534-5807(04)00058-9
- J. Aruga, K. Mizugishi, H. Koseki, K. Imai, R. Balling, T. Noda and K. Mikoshiba, “Zic1 Regulates the Patterning of Vertebral Arches in Cooperation with Gli3,” Mechanisms of Development, Vol. 89, No. 1-2, 1999, pp. 141- 150. doi:10.1016/S0925-4773(99)00220-8
- D. Krakow, S. P. Robertson, L. M. King, T. Morgan, E. T. Sebald, C. Bertolotto, S. Wachsmann-Hogiu, D. Acuna, S. S. Shapiro, T. Takafuta, S. Aftimos, C. A. Kim, H. Firth, C. E. Steiner, V. Cormier-Daire, A. Superti Furga, L. Bonafe, J. M. Graham, A. Grix, C. A. Bacino, J. Allanson, M. G. Bialer, R. S. Lachman, D. L. Rimoin and D. H. Cohn, “Mutations in the Gene Encoding Filamin B Disrupt Vertebral Segmentation, Joint Formation and Skeletogenesis,” Nature Genetics, Vol. 36, No. 4, 2004, pp. 405-410. doi:10.1038/ng1319
- B. Thomsen, P. Horn, F. Panitz, E. Bendixen, A. H. Petersen, L. E. Holm, V. H. Nielsen, J. S. Agerholm, J. Arnbjerg and C. Bendixen, “A Missense Mutation in the Bovine SLC35A3 Gene, Encoding a UDP-N-Acetylglucosamine Transporter, Causes Complex Vertebral Malformation,” Genome Research, Vol. 16, No. 1, 2006, pp. 97- 105. doi:10.1101/gr.3690506
- S. Yokoyama, Y. Ito, H. Ueno-Kudoh, H. Shimizu, K. Uchibe, S. Albini, K. Mitsuoka, S. Miyaki, M. Kiso, A. Nagai, T. Kikata, T. Osada, N. Fukuda, S. Yamashita, D. Harada, V. Mezzano, M. Kasai, P. L. Puri, Y. Hayashizaki, H. Okado, M. Hashimoto and H. Ashara, “A Systems Approach Reveals That the Myogenesis Genome Network Is Regulated by the Transcriptional Repressor RP58,” Developmental Cell, Vol. 17, No. 6, 2009, pp. 836-848. doi:10.1016/j.devcel.2009.10.011
- P. Soriano, “The PDGF Alpha Receptor Is Required for Neural Crest Cell Development and for Normal Patterning of the Somites,” Development, Vol. 124, No. 14, 1997, pp. 2691-2700.
- D. Chapman, A. Cooper-Morgan, Z. Harrelson and V. Papaioannou, “Critical Role for Tbx6 in Mesoderm Specification in the Mouse Embryo,” Mechanisms of Development, Vol. 120, No. 7, 2003, pp. 837-847. doi:10.1016/S0925-4773(03)00066-2
- C. Keegan, J. Hutz, T. Else, M. Adamska, S. Shah, A. Kent, J. Howes, W. Beamer and G. Hammer, “Urogenital and Caudal Dysgenesis in Adrenocortical Dysplasia (acd) Is Caused by a Splicing Mutation in a Novel Telomeric Regulator,” Human Molecular Genetics, Vol. 14, 2005, pp. 113-123. doi:10.1093/hmg/ddi011
- N. A. Quaderi, S. Schweiger, K. Gaudenz, B. Franco, E. I. Rugarli, W. Berger, G. J. Feldman, M. Volta, G. Andolfi, S. Gilgenkrantz, R. W. Marion, R. C. Hennekam, J. M. Optiz, M. Muenke, H. H. Ropers and A. Ballabio, “Opitz G/BBB Syndrome, a Defect of Midline Development, Is Due to Mutations in a New RING Finger Gene on Xp22,” Nature Genetics, Vol. 17, No. 3, 1997, pp. 285-291. doi:10.1038/ng1197-285

