Computational Molecular Bioscience
Vol.2 No.3(2012), Article ID:22488,4 pages DOI:10.4236/cmb.2012.23006
Amino Acid Biosynthesis and Proteolysis in Lactobacillus bulgaricus Revisited: A Genomic Comparison
1Graduate School of Bioresource Sciences, Nihon University, Fujisawa, Japan
2Division of Research and Development, Meiji Co., Ltd., Odawara, Japan
3Shanghai Institutes for Biological Science, Chinese Academy of Sciences, Shanghai, China
4Chinese National Human Genome Center at Shanghai, Shanghai, China
5WHO Collaborating Centre for Research in Human Reproduction, Shanghai, China
Email: *zai-si_ji@kta.biglobe.ne.jp, zhenghj@chgc.sh.cn
Received April 29, 2012; revised June 5, 2012; accepted June 15, 2012
Keywords: Amino Acid; Biosynthesis; Proteolysis; L. bulgaricus
ABSTRACT
The amino acid biosynthesis and proteolytic system of Lactobacillus bulgaricus (L. bulgaricus) is important for its growth in niche-specific environments, as well as for flavour formation in the food industry. Comparative analyses of 4 completed sequences of the L. bulgaricus strain genome on a genomic scale revealed that genes involved in amino acids synthesis were undergoing reductive evolution. However, the selected industrial strains, namely, L. bulgaricus 2038 and L. bulgaricus ND02, retained more complete genes in the amino acid synthesis and proteolytic system category than the laboratory strains, and have some unique genes and pathways for obtaining amino acids that enable these bacteria to adapt to their various environmental niches.
1. Introduction
The most important application of Lactobacillus bulgaricus (L. bulgaricus) is as a starter in the manufacture of various fermented dairy products. Its biochemical activity is not only capable of lactic acid production but is also responsible for protein hydrolysis and amino acid biosynthesis, which generates peptides and amino acids for bacterial growth and produces metabolites that contribute to flavor formation in fermented products [1]. The amino acid catabolism system of L. bulgaricus functions to balance the bacterium’s requirement for amino acids, and its proteolytic system plays a key role in its growth [2,3]. In particular, the functions of cell-wall-bound proteinases and peptidases are the most important functions for cell growth under conditions containing different types of nitrogen.
Several reports describing the proteolytic system of lactic acid bacterial (LAB) with respect to their biochemical and genetic aspects [4,6] have included little information specific to L. bulgaricus. However, putative genetic mechanisms underlying amino acid biosynthesis and proteolysis in this bacterial species have not been studied in detail. In the past few years, 4 L. bulgaricus genome strains (L. bulgaricus ATCC11842, L. bulgaricus ATCC BAA365, L. bulgaricus 2038, and L. bulgaricus ND02) have been completely sequenced; in addition, some strains have been incompletely sequenced to yield information regarding the proteolytic system, which has now allowed a thorough comparative analysis of their amino acid biosynthesis pathway and proteolytic systems on a genomic scale. Furthermore, the available genomic information could provide new insights into the genetic aspects of amino acid biosynthesis and proteolysis within L. bulgaricus through the identification of different genetic events at the genomic level. Comparative genomics has revealed some differences between the amino acid biosynthesis and proteolytic systems within Lactobacillus, the differences that are thought to reflect the various environmental niches that these bacteria occupy [7].
In a preliminary study, we described an in silico analysis of the L. bulgaricus 2038 amino acid biosynthesis and proteolytic system from its complete sequence, which will be publicly available in 2012 [8]. In this study, we describe an in-depth bioinformatics analysis in which we have systematically explored the diversity of the amino acid biosynthesis and proteolytic system in 4 completely sequenced L. bulgaricus strains. Based on our results, we have predicted horizontally unique genes in these 4 completely sequenced L. bulgaricus strains, with a focus on the genes required for bacterial growth, and also those that are niche-specific. Distinctions among the bacterial amino acid biosynthesis pathways, cell-wallbound proteinases and peptidases, peptide/amino acid transport systems, and intracellular proteinases and peptidases are described in detail as examples. Furthermore, results of comparative genomics analysis were used to explore the diversity of members of the proteolytic system in 4 L. bulgaricus strains by using pan-genome CGH analysis.
2. Computational Methods
2.1. Comparative Genomic Analyses
Complete genome sequences of L. bulgaricus (L. bulgaricus ATCC11842, L. bulgaricus ATCC BAA365, L. bulgaricus ND02, and L. bulgaricus 2038) were obtained from the NCBI microbial genome database (http://www.ncbi.nlm.nih.gov/genomes/MICROBES/microbial_taxtree.html). Comparative analysis was performed using progressive MAUVE [9]. Peptidase classification was performed using MEROPS Blast Server [10] with E value 0.0001.Metabolic pathway analysis was performed based on KEGG databse [11] through bi-directional best hit method. Extracellular and transmembrane proteins were determined by SignalP3.0 [12], ConPred II [13], and PSORTb v.2.0 [14]. COG assignment was performed using RPS-BLAST againt CDD (conserved domain database) [15] with E-value 1E-3. Protein homology was determined using BlastP with identity > 20% and length coverage > 30%, and alignment of protein sequences was performed using ClustalW.
2.2. Identification of Orthologous Groups
Orthologous groups in the 4 genomes were defined using the MBGD database [16]. The phylogenetic position of L. bulgaricus was determined on the basis of ortholog proteins. Concatenated protein sequences were first aligned using ClustalW, and the conserved alignment blocks were then extracted using the Gblocks program [17]. A maximum-likelihood tree was built using PHYML [18] with the following parameters: 100 replications for bootstrap analysis, “JTT” for the substitution model, “estimated” for the proportion of invariable sites, “estimated” for gamma distribution parameters, “4” for the number of substitution categories, “yes” to optimize tree topology, and “BIONJ” for starting tree(s).
3. Results
3.1. Genome Features of Complete Sequenced L. bulgaricus Genomes
Regarding the 4 completed genomes of L. bulgaricus, strains ATCC11842 and ATCC BAA365 are laboratory strains, and strains 2038 and ND02 are used as industrial strains. Compared to the other 3 strains having a genome size ranging from 1856 kb to 1872 kb with no plasmid, strain ND02 exhibited the greatest genome size at 2,125,753 bp, which was approximately 250 kb larger than the other genomes, and was also accompanied by a 6223-bp plasmid. Comparative genomic analysis showed that the 4 genomes are conserved in their genome
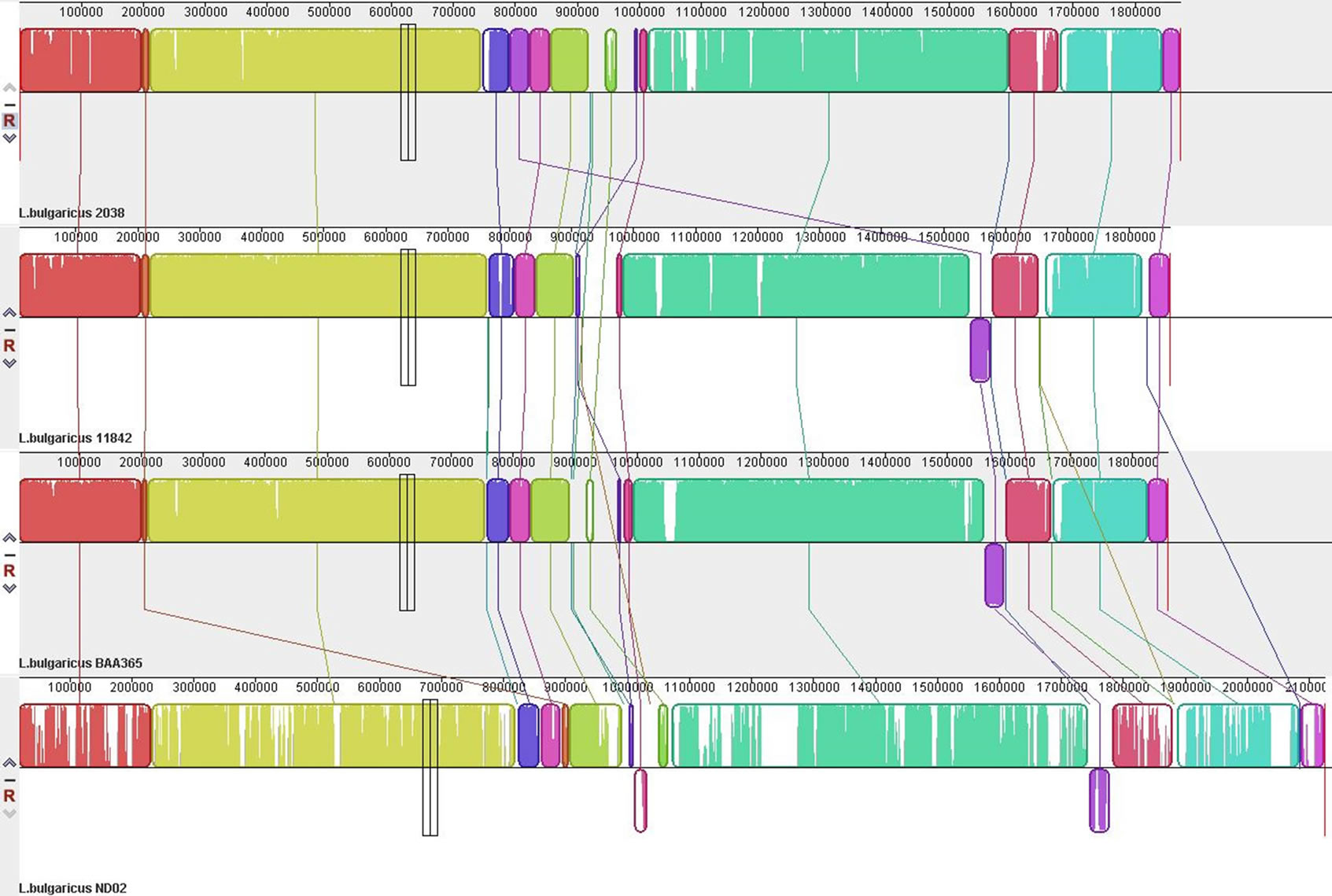
Figure 1. Alignment of the 4 L. bulgaricus complete genomes.
structure (Figure 1). There are a total of 1,466,078-bp sequences distributed in 451 blocks, which might be considered the “core genome” of L. bulgaricus. Meanwhile, 531,087-bp sequences were deemed strain ND02- unique; 136,451-bp sequences as strain 2038-unique; 122,623-bp sequences as strain ATCC 11842-unique; and 118,323-bp sequences as strain ATCC BAA365-unique.
Detailed analysis of the enzymes involved in de novo amino acid biosynthesis revealed 39 proteins in strain 2038, 38 proteins in strain ND02, 21 proteins in strain ATCC BAA365, and 18 proteins in strain ATCC 11842 (Table S1). Regarding the proteolytic systems compared among these 4 genomes, strain ND02 possessed 81 proteases and peptidases, while the number in strains 2038, ATCC 11842, and ATCC BAA365 were 72, 64, and 67, respectively (Table S2, S3). In addition, no unique protease/peptidases in each strain were found according to the cluster of ortholog (COG) among these 4 genomes, although the number of proteases/peptidases varied among different strains. For peptide/amino acid transport systems, strain 2038 possessed the highest number, which encoded 90 proteins involved in this system, while the strains ND02, ATCC BAA365, and ATCC 11842 encoded 75, 61, and 60 proteins, respectively. The highest number of proteins participating in peptide transporters in strain 2038 was mainly due to 24 genes encoding ABC-type oligopeptide transport system substratebinding protein, which had only 18 genes in ND02, 6 genes in ATCC 11842, and 5 genes in ATCC BAA365 strains.
3.2. Gene Gain or Loss in These L. bulgaricus Strains
A phylogenetic tree based on proteins of amino acid synthesis and proteolysis systems was constructed (Figure 2) for these 4 strains. The genes involved in these 2 systems might have evolved with adaption to different growth environments and passed through different selection processes, and may have played a key role in the growth of the L. bulgaricus strain. Strain ND02 was located on a separate branch compared to the other 3 strains, coinciding with its larger genome size and its plasmid. Strain 2038 retained more genes related to amino acid synthesis and proteolysis systems from a common ancestor genome compared to the other 2 laboratory strains.
A total of 1682 ortholog groups were identified within these 4 L. bulgaricus strains, in which 1232 ortholog groups existed in all 4 genomes and might be considered the “core genes” of L. bulgaricus. Another 268 ortholog groups were revealed in strain ND02 and in at least 1 of the other 3 strains that might be considered “dispensable” genes; these may have existed in the ancestor genome, but could have been subsequently lost during the divergence process that produced strain 2038 and/or the other 2 laboratory strains. Meanwhile, the ortholog groups among these 4 strains were analyzed, and the genes gained or lost during the evolutionary process of each strain were identified. Strain 2038 lost 67 genes after it diverged from strain ND02, while strain ATCC11842 lost over 200 genes and ATCC BAA365 lost 150 genes. Among the 67 genes lost in strain 2038, only 2 encoded for proteases, while the others had nothing to do with amino acid synthesis or proteolysis systems. However, for the 2 laboratory strains, 15 genes involved in amino acid synthesis, 12 genes encoding proteases, and 16 genes participating in transport systems were lost (Table S4).
3.3. Unique Genes and Pathways Involved in Amino Acid Synthesis in L. bulgaricus
The distribution of enzymes involved in the amino acid biosynthesis pathway in these 4 completely sequenced L. bulgaricus genomes is given in Table S1. Based on the analysis of de novo amino acid synthesis ability (Table 1),
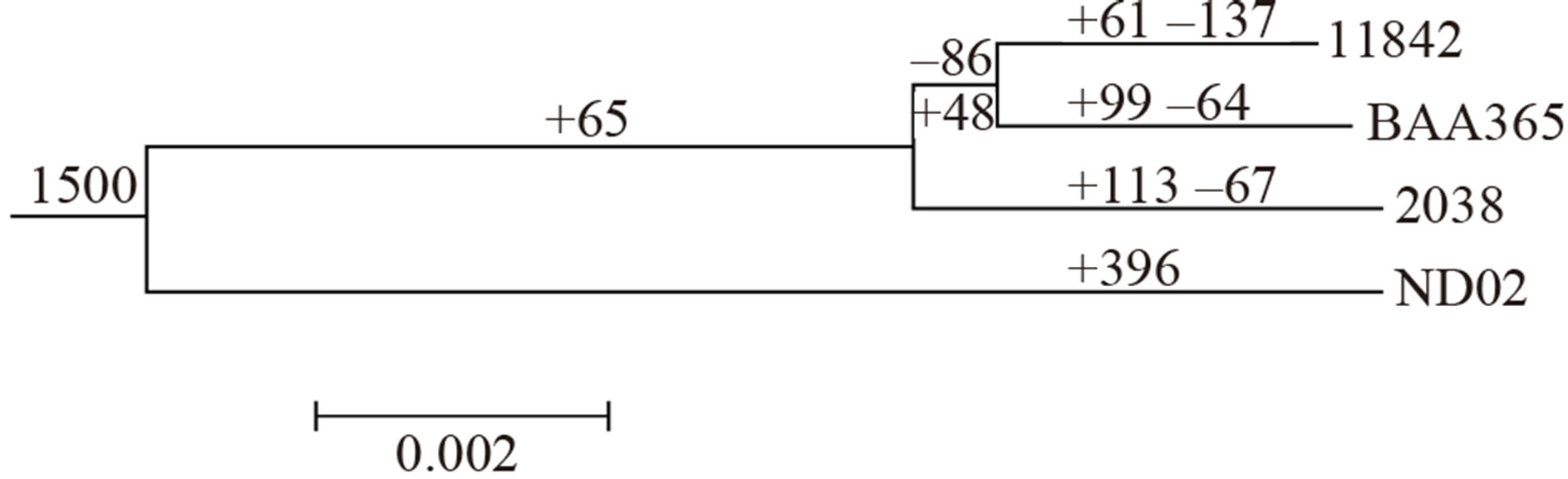
Figure 2. Phylogenetic tree showing evolutionary divergence in the 4 L. bulgaricus genomes based on the results of analyses of amino acid synthesis and paroteolysis systems. The number on each branch represented gene family number gained (+) or lost (−) during evolution.
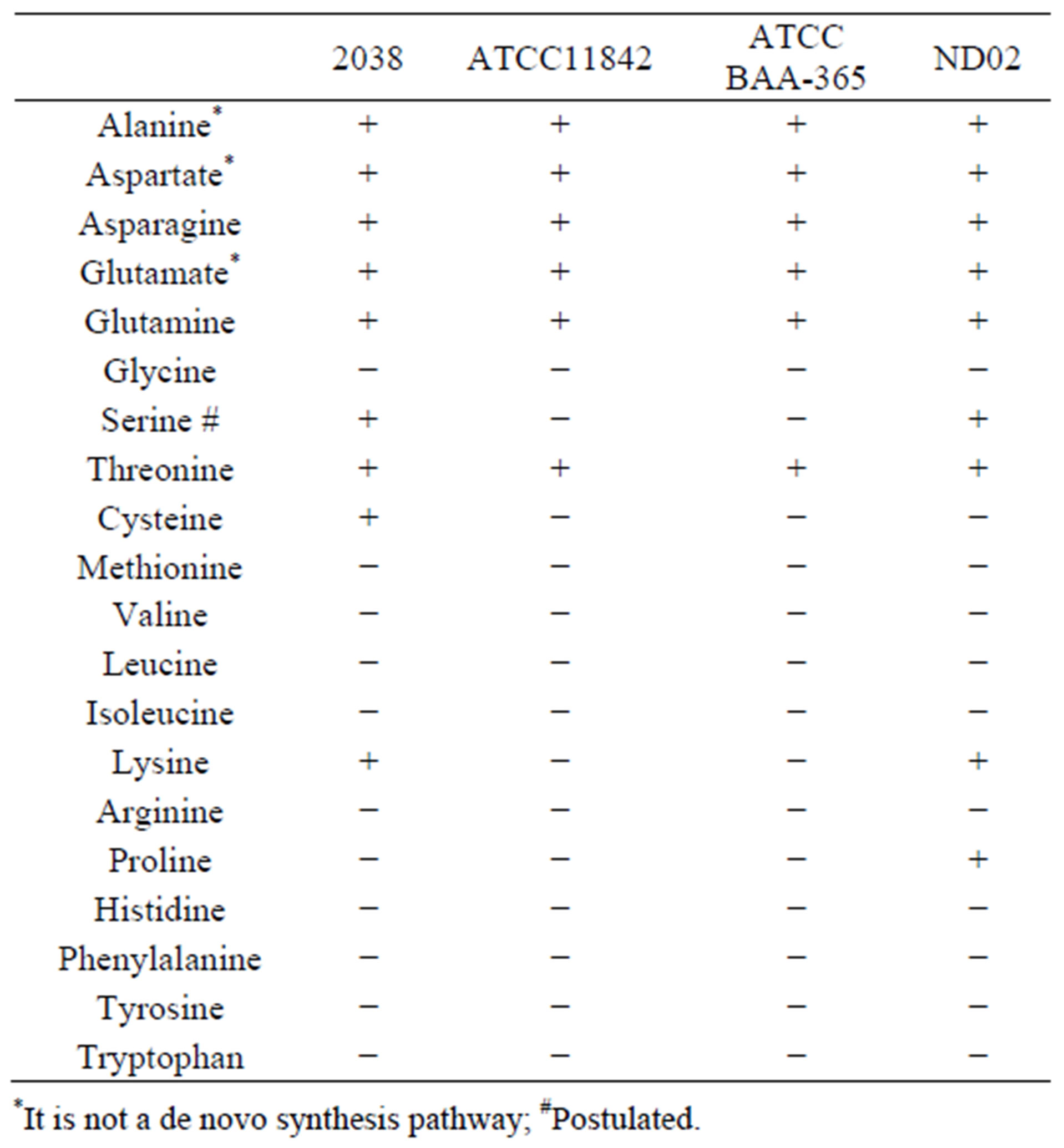
Table 1. Statistics of amino acid synthesis in L. bulgaricus genomes.
threonine, asparagine, and glutamine could be synthesized from l-aspartate by all 4 of these L. bulgaricus strains. Meanwhile, alanine could be converted from cysteine and glutamate could be transformed from aspartate, and vice versa (Table S1).
Proline could be biosynthesized from glutamate only in L. bulgaricus ND02 via 3 enzyme-catalyzed reactions. On the other hand, in the other 3 L. bulgaricus strains, all 3 genes encoding for the 3 enzymes were lost, except for gamma-glutamyl kinase (EC: 2.7.2.11), which was encoded by strain 2038 (LBU0712). Cysteine can only be synthesized from serine through 2 enzyme-catalyzed reactions in L. bulgaricus 2038. The first step is mediated by serine O-acetyltransferase (cysE, EC: 2.3.1.30, encoded by LBU1138), which was lost in the other 3 L. bulgaricus strains. The second step is catalyzed by cysteine synthase A (cysK, EC: 2.5.1.47, encoded by LBU1136 or LBU1253).
Lysine could be synthesized in the 2 industrial strains ND02 and 2038 by 7 genes encoding enzymes from dihydrodipicolinate synthase (EC: 4.2.1.52) to diaminopimelate epimerase (EC: 5.1.1.7); the genes are clustered together, and are missing in 2 laboratory strains. Most likely, an acetyl rather than a succinyl-catalyzed reaction took place with regard to enzyme DapD (EC: 2.3.1.117). The other 2 genes encoding Asd (EC: 1.2.1.11) and LysA (EC: 4.1.1.20) were positioned outside the cluster.
Another pathway unique to both industrial strains is serine biosynthesis. This pathway, which starts from 3-phosphoglycerate, includes 3 enzymes; genes encoding the first 2 enzymes, namely, phosphoglycerate dehydrogenase (serA, EC: 1.1.1.95) and phosphoserine aminotransferase (serC, EC: 2.6.1.52), were found in both strains (Table S1). Although the gene encoding the latter enzyme phosphoserine phosphatase (serB, EC: 3.1.3.3) was not found in both genomes, 2 homologous genes (LBU0835/LDBND_0867 and LBU0857/LDBND_0902) were found that might encode proteins performing a function similar to phosphoserine phosphatase.
A unique pathway responsible for amino acid synthesis exists in L. bulgaricus; this pathway is different from that found in any other microorganism: alanine is normally produced from pyruvate through alanine transaminase (alt, EC: 2.6.1.2), alanine dehydrogenase (ald, EC: 1.4.1.1), or aspartate 4-decarboxylase (asdA, EC: 4.1.1.12), but no such genes or domains are found in L. bulgaricus. There are 2 cysteine desulfurase genes (nifS, EC: 2.8.1.7) in all 4 L. bulgaricus genomes, suggesting that alanine might be produced from cysteine through a reaction catalyzed by cysteine desulfurase.
3.4. Sequence Comparison to Distinguish Cell-Wall-Bound Proteases and Peptidases
Six extracellular proteases are conserved among all 4 L. bulgaricus strains, including 2 serine proteases and 4 metalloproteases; meanwhile, both strains BAA365 and ND02 encode another unique membrane protease (Table S5). The protein sequences of the most important proteinase in L. bulgaricus proteolysis, namely, PrtB (EC: 3.4.21.83), are highly conserved among the 4 strains. This protein also has identical amino acids around the catalytic sites (Asp-30, His-94, Asn-189, Ser-425) and substrate-binding sites (Gly-131, Val-159, Gly-728, Thr-729). However, PrtB lacked a fragment of 125 amino acids (from residue 1450 to 1574 in mature protein) at the C-terminus in L. bulgaricus 2038 and ATCC BAA365, which is rich in Asp and Lys and located 100 amino acids ahead of the sorting signal (LPKKT) (Figure 3). This region contains an alpha-helix structure of H domain and the first 45 amino acids of transwall domain (W domain). As is known that H domain is able to position the N-terminal of PrtB outside the cell wall, and W domain spans the cell wall [19]. The loss of this region might affect the attachment to cell wall and folding of PrtB, thus changing the cleavage pattern on substrate.
The maturation of PrtB depends upon PrtM, which could not be identified in the 4 L. bulgaricus strains. Based on a search for homologs, the foldase protein PrsA (EC: 5.2.1.8) was identified in all 4 strains—PrsA is known to be an extracellular chaperon involved in extracellular protease maturation, as well as the folding and stability of subtilisins in Bacillus subtilis [20]. In comparison with known PrtM proteins, PrsA from the 4 L. bulgaricus strains was highly homologous to PrtM (Figure 4). The conserved domain of PrtM and PrsA had little variation, supporting the possibility that PrsA take up the role of PrtM.
Two extracellular peptidases involved in peptide degradation varied greatly among the 4 L. bulgaricus strains. The endopeptidase EnlA, encoded by LBU1040, had no ortholog in strain ATCC BAA365, and the dipeptidase PepD4, EC: 3.4.-.-, encoded by LBU1705, had no ortholog in ATCC 11842. Through protein sequence alignment, the strains 2038, ATCC 11842, and ND02 showed a difference at the C-terminus of EnlA, and no domain was observed in this variable region. For PepD4, strain ND02 lost the largest proportion of the 396-amino acid-long domain PF03577 (from the 24th to the 422nd cordon in LBU1705); thus, its peptidase activity might have also been lost, although it encoded an extracellular homolog of PepD4 (Figure 5).
A cell surface housekeeping protease, HtrA (EC: 3.4. 21.-), was identified in the 4 L. bulgaricus strains, and is known to be involved in protein maturation and turnover [21]. A comparison of the HtrA proteins within all 4 L. bulgaricus strains revealed high similarities among the groups (Figure 6).

Figure 3. Sequence alignment of mature PrtB. The red star indicates conserved catalytic sites, blue star indicates substratebinding site and black star indicates the sorting signal.
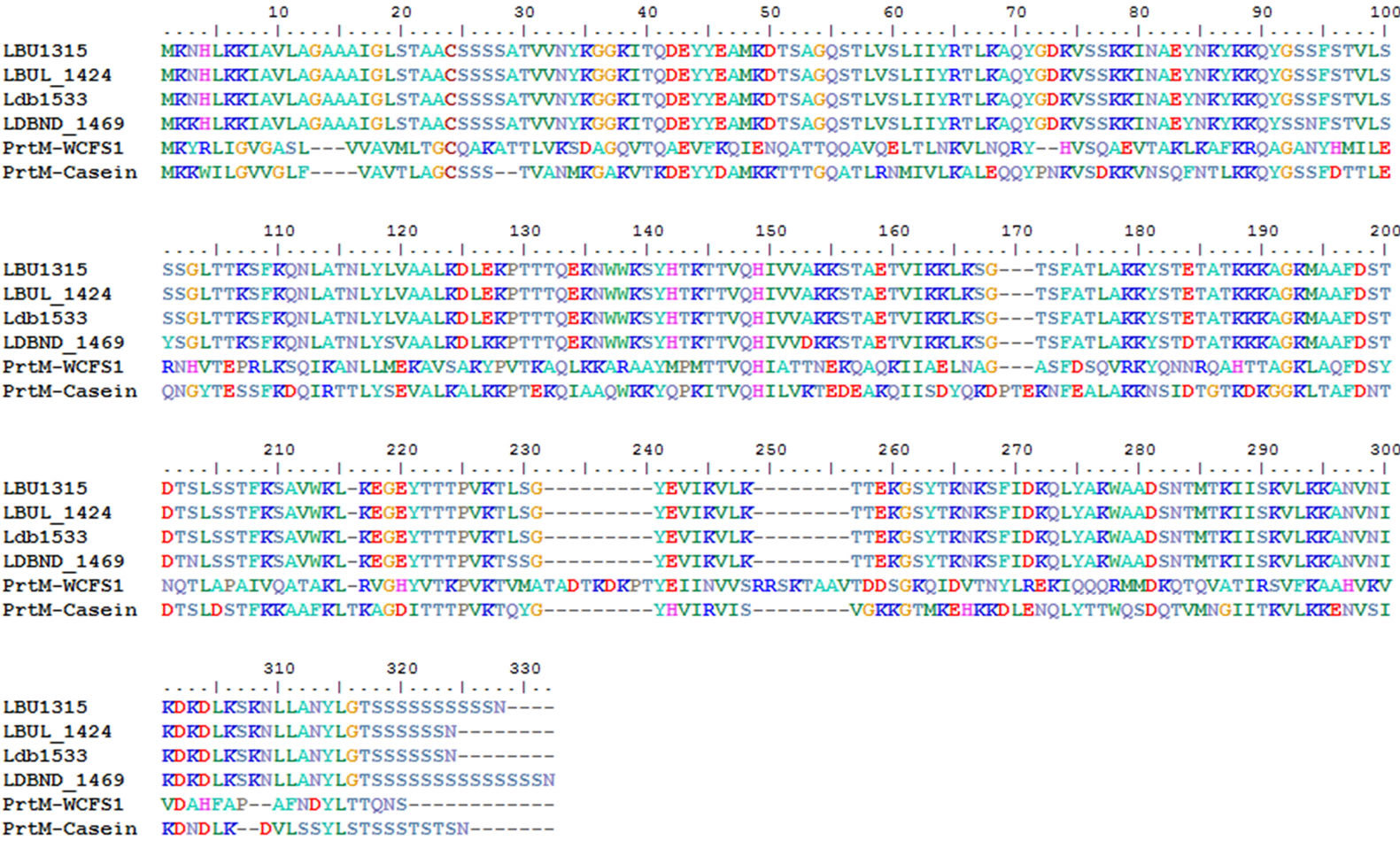
Figure 4. Alignment and domain comparisons in PrtM and PrsA.

Figure 5. Alignment of the extracellular peptidase En1A (A) and PepD4 (B).
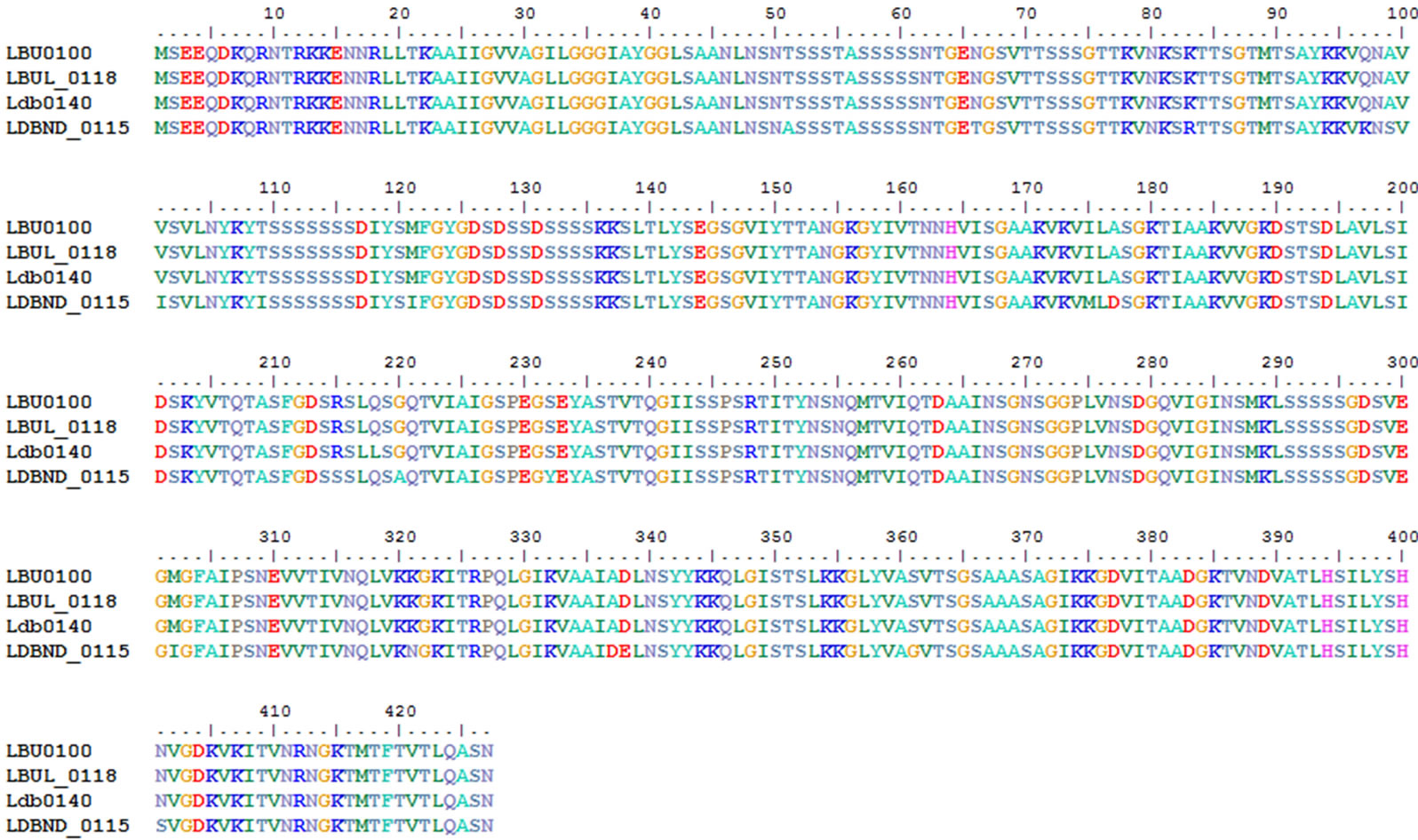
Figure 6. Alignment and domain comparisons in HtrA.
3.5. The Distribution of Intracellular Peptidase in Sequenced L. bulgaricus Genomes
Two intracellular proteases were highly conserved, but the intracellular peptidases showed some differences among these 4 L. bulgaricus strains. Thirty-three intracellular peptidases were found in L. bulgaricus strain 2038. Through comparison, we found that all intracellular proase/peptidases of strain ATCC11842 and ATCC BAA365 had homologs in strain 2038, while strain ND02 encoded 3 unique peptidases, including 2 serine peptidases (LDBND_1003 and LDBND_1120), and 1 metallopeptidase (LDBND_179). Furthermore, strain 2038 encoded 4 peptidases whose homolog was found only in 1 of the other 3 strains, including 1 aminopeptidase2 carboxypeptidases, and 1 endopeptidase (Table S6). Among them, the homolog of d-aminopeptidase DppA (encoded by LBU0520) was only found in strain ATCC 11842, the homolog of metal-dependent amidase/aminoacylase/carboxypeptidase (encoded by LBU0898) was only found in strain ATCC BAA365, and an amino acid amidohydrolase (encoded by LBU0934) and a metalloendopeptidase (encoded by LBU1255) had orthologs only in strain ND02.
Two cysteine aminopeptidase genes (PepG1-LBU0223 and PepG2-LBU0224) clustered together to supply all the L. bulgaricus strains with cysteine. Alignment studies showed that the 2 peptidases were highly conserved (Figure 7).
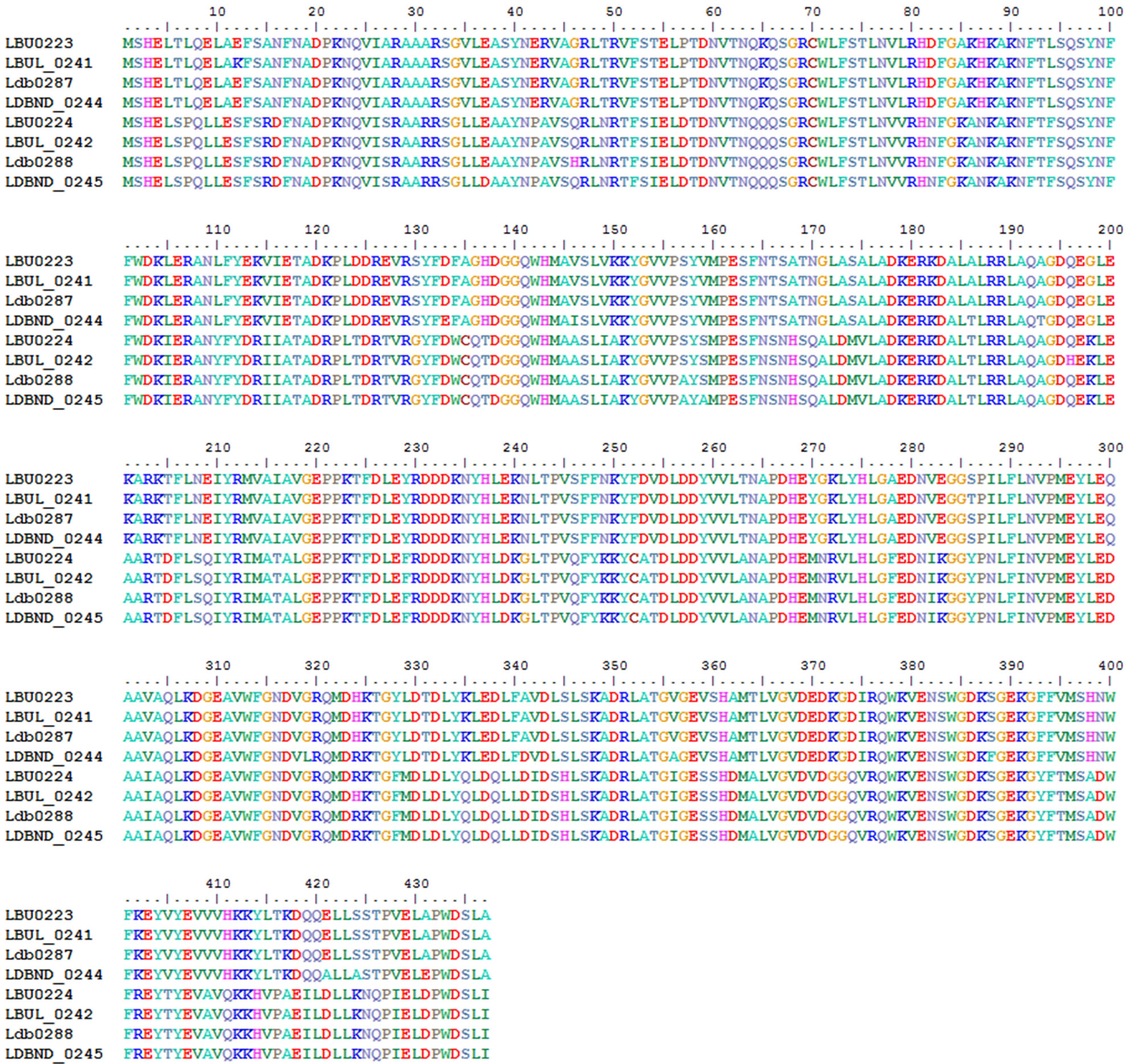
Figure 7. Alignment of cysteine aminopeptidase in the 4 L. bulgaricus strains.
3.6. Distribution of the Transport System in Sequenced L. bulgaricus Genomes
All 4 L. bulgaricus strains possess 2 complete oligoepde transport (Opp) systems (OppABCDF type and oppDFBCA type) [22], each with a very different number and order for substrate-binding proteins (OppA). With respect to the ABC-type transporters and permeases for single amino acids, the strains showed significant differences (Table S7).
Whereas 90 genes encoding amino acid/oligopeptide transporters were found in strain 2038, we found 75, 60, and 61 transporter genes, respectively, in the strains ND02, ATCC 11842, and ATCC BAA365 (Table S3). This large variation was mainly because of the number of OppA proteins—there were 25 of these in strain 2038, compared to 19 in ND02, and only 6 in ATCC 11842 and 7 in ATCC BAA365 (Table S7). In contrast, strains ATCC 11842 and ATCC BAA365 had no unique OppA proteins, while 5 were found in strain ND02, and 6 in strain 2038.
In addition to Opp. systems, 14 amino acid permeases were identified in the 4 L. bulgaricus strain genomes (Table S7). Five of the amino acid permeases were conserved, and might play roles in importing essential amino acids for L. bulgaricus strains. Three amino acid permeases (AppA2, LysP2, and ArcD3) were found only in the 2 industrial strains. Meanwhile, there were 2 unique amino acid permeases (encoded by LBU0206 and LBU- 0209) in strain 2038 and 1 in strain ND02 (encoded by LDBND1404). No unique amino acid permease was found in any of the laboratory strains, possibly illustrating that the laboratory strains had undergone a more extensive reductive evolution.
4. Discussion
In this study, we performed a systematic genome-wide analysis of all the proteins involved in amino acid biosynthesis and proteolysis, from 4 completely sequenced L. bulgaricus genomes, including the strains ATCC 11842, ATCC BAA-365, ND02, and 2038. Comparative genomics analysis was conducted to distinguish various subgroups within a protein superfamily, allowing for a highly improved annotation of genes, and clarification of any inconsistent annotations. This information could be used to predict the amino acid catabolism potential for all L. bulgaricus strains.
Two pathways have been identified in bacteria to assimilate ammonia into glutamate: 1 is the glutamate dehydrogenase (GDH, EC: 1.4.1.2) pathway, and another is via the glutamine synthase (GS, EC: 6.3.1.2)-glutamate synthase (GOGAT, EC: 1.4.1.13/14) cycle [23]. Both pathways form 1 molecule of glutamate from 1 molecule of ammonia (HN3) and 1 molecule of 2-oxoglutarate. However, no genes encoding for GDH or GOGAT ware identified in these 4 completely sequenced L. bulgaricus genomes. Similar to how l-aspartate is synthesized from oxaloacetate and glutamate through the catalysis of aspartate aminotransferase (EC: 2.6.1.1), glutamate could be produced in the same manner from 2-oxoglutarate and l-aspartate. We found 2 genes encoding proteins homologous to aspartate aminotransferase in the 4 L. bulgaricus strain genomes (Table S1). These results suggest that aspartate aminotransferase catalyzes the formation of both glutamate and aspartate in L. bulgaricus.
All the L. bulgaricus strains similarly possessed limited anabolic biosynthesis capabilities because of a partial TCA cycle that contained only fumarate reductase (EC: 1.3.99.1) and fumarate hydratase (EC: 4.2.1.2). Six kinds of amino acids (aspartate, asparagine, glutamine, glutamate, threonine, and alanine) could be biosynthesized in all 4 strains. According to the results from the annotation studies, glutamate, aspartate, glutamine, and alanine were related to additional amino acid biosynthesis, nucleoside metabolism, and cell-wall formation. This suggests that L. bulgaricus took advantage of the nitrogen-rich environment when they lost the capacity for ammonium assimilation and glutamate biosynthesis.
Only the industrial strains 2038 and ND02 had the ability to biosynthesize lysine and serine by using a separate pathway for each amino acid. Meanwhile cysteine could only be synthesized by 2038 and proline only by ND02 through unique pathways for each. In addition to losing genes, each strain evolved new genes or retained ancestor genes after divergence. Among the 396 unique genes for strain ND02, 24 (6%) were associated with the amino acid synthesis/proteolysis system, whereas in 113 unique genes for strain 2038, 10 (8.8%) were inlved in its amino acid synthesis/ roteolysis system. Except for the 2 ATCC strains, only 5 unique genes encoding proteases or transporters were found in all the strains studied (Table S8). Based on the presence of genes encoding protease and transport systems, the 2 industrial stains were more capable of utilizing extrinsic resources than the 2 laboratory strains, and were likely influenced by industrial cultivation [2].
For the 4 genomes of the L. bulgaricus strains, the genes encoded a relatively lower number and variety of amino acid biosynthesis components, which lost some biosynthetic capability, as the strains continued to develop in different environments. Protein homolog analysis showed that only strains ND02 and 2038 had unique genes that are involved in amino acid synthesis in the 4 L. bulgaricus strains (Table S4). Strain ND02 had 6 genes encoding for proline synthesis (LDBND_0093, LDBND _0094, LDBND_0755, LDBND_0756, LDBND_0788, and LDBND_1721), which was the highest number for this function among the strains studied. Strain 2038 possessed a unique gene, LBU1138, which encoded serine ο- acetyltransferase, and therefore, endowed this strain with the ability to synthesize cysteine. Although strain ND02 possessed the biggest genome size and the largest unique region, it did not show significantly improved amino acid catabolism and proteolysis compared to strain 2038.
Strain 2038 had a growth advantage relative to the other strains because it could potentially synthesize cysteine, which is difficult to obtain from exogenous casein and can be used as a precursor of alanine. The expression of 2 concatenated cysteine aminopeptidase genes might increase to satisfy the demand for cysteine in L. bulgaricus cells or to carry out the same function under different conditions. Strain ND02 could synthesize proline, which might afford it a growth-related advantage over the other strains. These features explain why strains 2038 and ND02 were chosen as the industrial strains; in addition, they possess more genes associated with nitrogen utilizetion compared to the other strains.
Evolutionary differences between the 2 industrial strains, as illustrated in the phylogenetic tree that was generated from analyzing amino acid synthesis and proteolysis genes, could be attributed to the fact that each industrial strain was isolated from a different geographical area. Therefore, differences between using cow milk from Europe and sheep milk from North Asia, and lower cysteine concentrations in cow milk compared to sheep milk, and differences between these 2 areas feeding environments, may have all contributed to the observed evolutionary divergence between the 2 strains.
Genomic sequence analysis revealed the presence of at least 45 genes encoding putative proteases or peptidases. From this group, the most significant among the genes encoding cell surface proteases is PrtB, which is located downstream of the lac operon in L. bulgaricus 2038 [24]. A comparison of PrtB among the 4 complete L. bulgaricus sequences showed that the protease catalytic sites and substrate-binding sites are conserved, while a 125-amino acid-long deletion in C-terminal was revealed in L. bulgaricus 2038 and might affect the folding of the enzyme. These results imply that different L. bulgaricus strains might have different substrate specificities and release different peptides.
The foldase protein PrsA was identified in the genomes of all 4 L. bulgaricus strains. Because this protein was very highly homologous to PrtM and had the same EC number, and no PrtM was found in L. bulgaricus until now, we suggest that it may play role in the maturation of PrtB instead of PrtM; this may explain the maturation and turnover rates of PrtB in all L. bulgaricus strains. The cell surface housekeeping protease HtrA was also identified in all 4 L. bulgaricus strain genomes. Considing that only 2 cell surface peptidases were identified in all 4 strains, it is reasonable to speculate that these 2 cell surface peptidases may perform a single function.
Two putative cell surface peptidases that are not heatshock proteinases—the membrane protein related to metalloendopeptidase EnlA and the dipeptidase PepD4— are either missing or were found as a pseudo gene either in strain ATCC BAA-365, or in strains ATCC 11842 and ND02 without a signal peptide in their C-termini. Only L. bulgaricus 2038 had these 2 completed genes with a signal. Considering that some intracellular peptidases were identified only in strain 2038, L. bulgaricus 2038 might potentially have a more powerful exterior protein degradation capability than other L. bulgaricus strains, which could be advantageous, as it would produce more free amino acids than the other strains. These amino acids could then be transported into the cell via 8 ATP-binding ABC-type amino acid transport systems or at least 6 permeases.
Two distinct opp operons were associated with multiple OppA proteins in the 4 L. bulgaricus strain genomes, but these 2 opp operons in strain 2038 are very interesting in their distinct operon structures; PepG1 and PepG2 (LBU0223 and LBU0224) encoding aminopeptidases are located directly adjacent to the OppF gene. There are also more OppA genes existing in strain 2038 compared to the other 3 strains. This might explain the important role of the Opp system in this industrial strain; different OppA proteins are used for different oligopeptides, especially, in different environments. Several peptide transporters or peptidases fall into larger protein super families: 1) aminopeptidase PepC and PepG belonging to the MEROPS peptidase family C1_B, 2) aminopeptidase PepI and PepL belonging to MEROPS family S33, and 3) aminopeptidase PepM together with dipeptidase PepQ and PepZ belonging to MAROPS family M24.
REFERENCES
- J. E. Christensen, E. G. Dudley, et al., “Peptidases and Amino Acid Catabolism in Lactic Acid Bacteria,” Antonie Van Leeuwenhoek, Vol. 76, No. 1-4, 1999, pp. 217- 246. doi:10.1023/A:1002001919720
- P. Hao, H. Zheng, et al., “Complete Sequencing and Pan-Genomic Analysis of Lactobacillus delbrueckii subsp. bulgaricus Reveal Its Genetic Basis for Industrial Yogurt Production,” PLOS One, Vol. 6, No. 1, 2011, p. e15964. doi:10.1371/journal.pone.0015964
- M. van de Guchte, S. Penaud, et al., “The Complete Genome Sequence of Lactobacillus bulgaricus Reveals Extensive and Ongoing Reductive Evolution,” Proceedings of the National Academy of Sciences of the United States of America, Vol. 103, No. 24, 2006, pp. 9274-9279. doi:10.1073/pnas.0603024103
- M. Liu, J. R. Bayjanov, et al., “The Proteolytic System of Lactic Acid Bacteria Revisited: A Genomic Comparison,” BMC Genomics, Vol. 11, No.1, 2010, p. 36. doi:10.1186/1471-2164-11-36
- M. Liu, A. Nauta, et al., “Comparative Genomics of Enzymes in Flavor-Forming Pathways from Amino Acids in Lactic Acid Bacteria,” Applied and Environmental Microbiology, Vol. 74, No. 15, 2008, pp. 4590-4600. doi:10.1128/AEM.00150-08
- M. Zhou, D. Theunissen, et al., “LAB-Secretome: A Genome-Scale Comparative Analysis of the Predicted Extracellular and Surface-Associated Proteins of Lactic Acid Bacteria,” BMC Genomics, Vol. 11, No. 1, 2010, p. 651. doi:10.1186/1471-2164-11-651
- J. Boekhorst, R. J. Siezen, et al., “The Complete Genomes of Lactobacillus plantarum and Lactobacillus johnsonii Reveal Extensive Differences in Chromosome Organization and Gene Content,” Microbiology, Vol. 150, No. 11, 2004, pp. 3601-3611. doi:10.1099/mic.0.27392-0
- H.-J. Zheng, E-N. Liu, et al., “In Silico Analysis of Amino Acid Biosynthesis and Proteolysis in Lactobacillus delbrueckii subsp. bulgaricus 2038 and the Implications for Bovine Milk Fermentation,” Biotechnology Letters, Vol. 34, No. 8, 2012, pp. 1545-1551. doi:10.1007/s10529-012-1006-4
- A. E. Darling, B. Mau, et al., “Progressivemauve: Multiple Genome Alignment with Gene Gain, Loss and Rearrangement,” Plos One, Vol. 5, No. 6, 2010, p. e11147. doi:10.1371/journal.pone.0011147
- N. D. Rawlings, A. J. Barrett, et al., “MEROPS: The Database of Proteolytic Enzymes, Their Substrates and Inhibitors,” Nucleic Acids Research, Vol. 40, D. 1, 2012, pp. D343-D350. doi:10.1093/nar/gkr987
- M. Kanehisa, S. Goto, et al., “The KEGG Resource for Deciphering the Genome,” Nucleic Acids Research, Vol. 32, Suppl. 1, 2004, D277-D280. doi:10.1093/nar/gkh063
- J. D. Bendtsen, H. Nielsen, et al., “Improved Prediction of Signal Peptides: SignalP 3.0,” Journal of Molecular Biology, Vol. 340, No. 4, 2004, pp. 783-795. doi:10.1016/j.jmb.2004.05.028
- M. Arai, H. Mitsuke, et al., “ConPred II: A Consensus Prediction Method for Obtaining Transmembrane Topology Models with High Reliability,” Nucleic Acids Research, Vol. 32, Suppl. 2, 2004, pp. W390-W393. doi:10.1093/nar/gkh380
- J. L. Gardy, C. Spencer, et al., “PSORT-B: Improving Protein Subcellular Localization Prediction for GramNegative Bacteria,” Nucleic Acids Research, Vol. 31, No. 13, 2003, pp. 3613-3617. doi:10.1093/nar/gkg602
- A. Marchler-Bauer, S. Lu, J. B. Anderson, et al., “CDD: A Conserved Domain Database for the Functional Annotation of Proteins,” Nucleic Acids Research, Vol. 39, Suppl. 1, 2011, pp. D225-D229. doi:10.1093/nar/gkq1189
- I. Uchiyama, T. Higuchi, et al., “MBGD Update 2010: Toward a Comprehensive Resource for exploring Microbial Genome Diversity,” Nucleic Acids Research, Vol. 38, Suppl. 1, 2010, pp. D361-365. doi:10.1093/nar/gkp948
- G. Talavera and J. Castresana, “Improvement of Phylogenies after Removing Divergent and Ambiguously Aligned Blocks from Protein Sequence Alignments,” Systematic Biology, Vol. 56, No. 4, 2007, pp. 564-577. doi:10.1080/10635150701472164
- S. Guindon, J. F. Dufayard, et al., “New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0,” Systematic Biology, Vol. 59, No. 3, 2010, pp. 307-321. doi:10.1093/sysbio/syq010
- J. E. Germond, M. Delley, et al., “Determination of the Domain of the Lactobacillus delbrueckii subsp. Bulgaricus Cell Surface Proteinase PrtB Involved in Attachment to the Cell Wall after Heterologous Expression of the PrtB Gene in Lactococcus lactis,” Applied Environmental Microbiology, Vol. 69, No. 6, 2003, pp. 3377-3384. doi:10.1128/AEM.69.6.3377-3384.2003
- M. Jacobs, J. B. Andersen, et al., “Bacillus subtilis PrsA Is Required in Vivo as an Extracytoplasmic Chaperone for Secretion of Active Enzymes Synthesized either with or without Pro-Sequences,” Molecular Microbiology, Vol. 8, No. 5, 1993, pp. 957-966. doi:10.1111/j.1365-2958.1993.tb01640.x
- N. Vermeulen, M. Pavlovic, et al., “Functional Characterization of the Proteolytic System of Lactobacillus sanfranciscensis DSM 20451T during Growth in Sourdough,” Applied Environmental Microbiology, Vol. 71, No. 10, 2005, pp. 6260-6266. doi:10.1128/AEM.71.10.6260-6266.2005
- S. Tynkkynen, G. Buist, et al., “Genetic and Biochemical Characterization of the Oligopeptide Transport System of Lactococcus lactis,” Journal of Bacteriology, Vol. 175, No. 23, 1993, pp. 7523-7532.
- M. Romero, S. Guzman-Leon, et al., “Pathways for Glutamate Biosynthesis in the Yeast Kluyveromyces lactis,” Microbiology, Vol. 146, No. 1, 2000, pp. 239-245.
- C. Gilbert, D. Atlan, et al., “A New Cell Surface Proteinase: Sequencing and Analysis of the PrtB Gene from Lactobacillus delbruekii subsp. bulgaricus,” Journal of Bacteriology, Vol. 178, No. 11, 1996, pp. 3059-3065.
Appendix
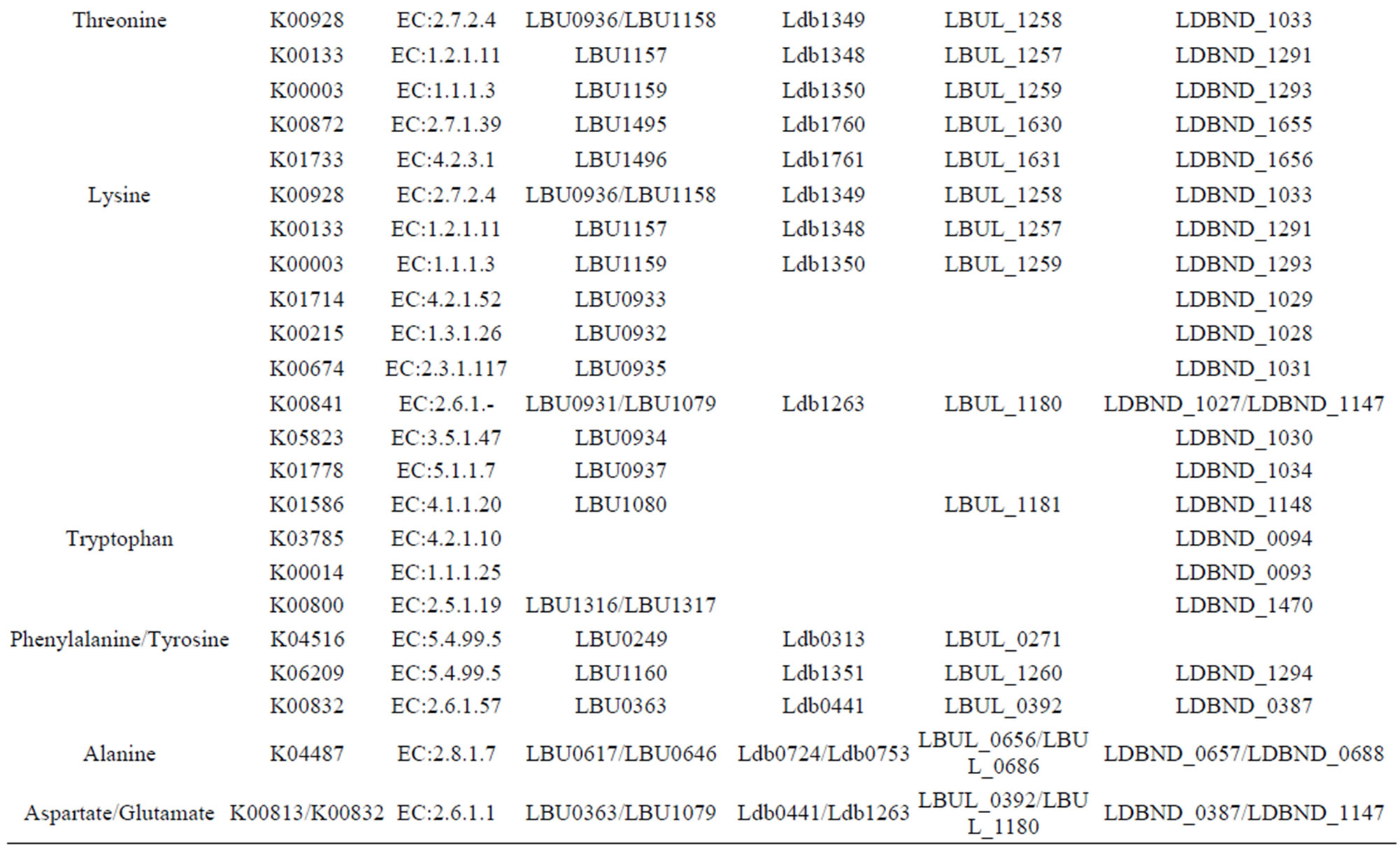

Table S1. Genes involved in amino acid biosynthesis for each L. bulgaricus strain.
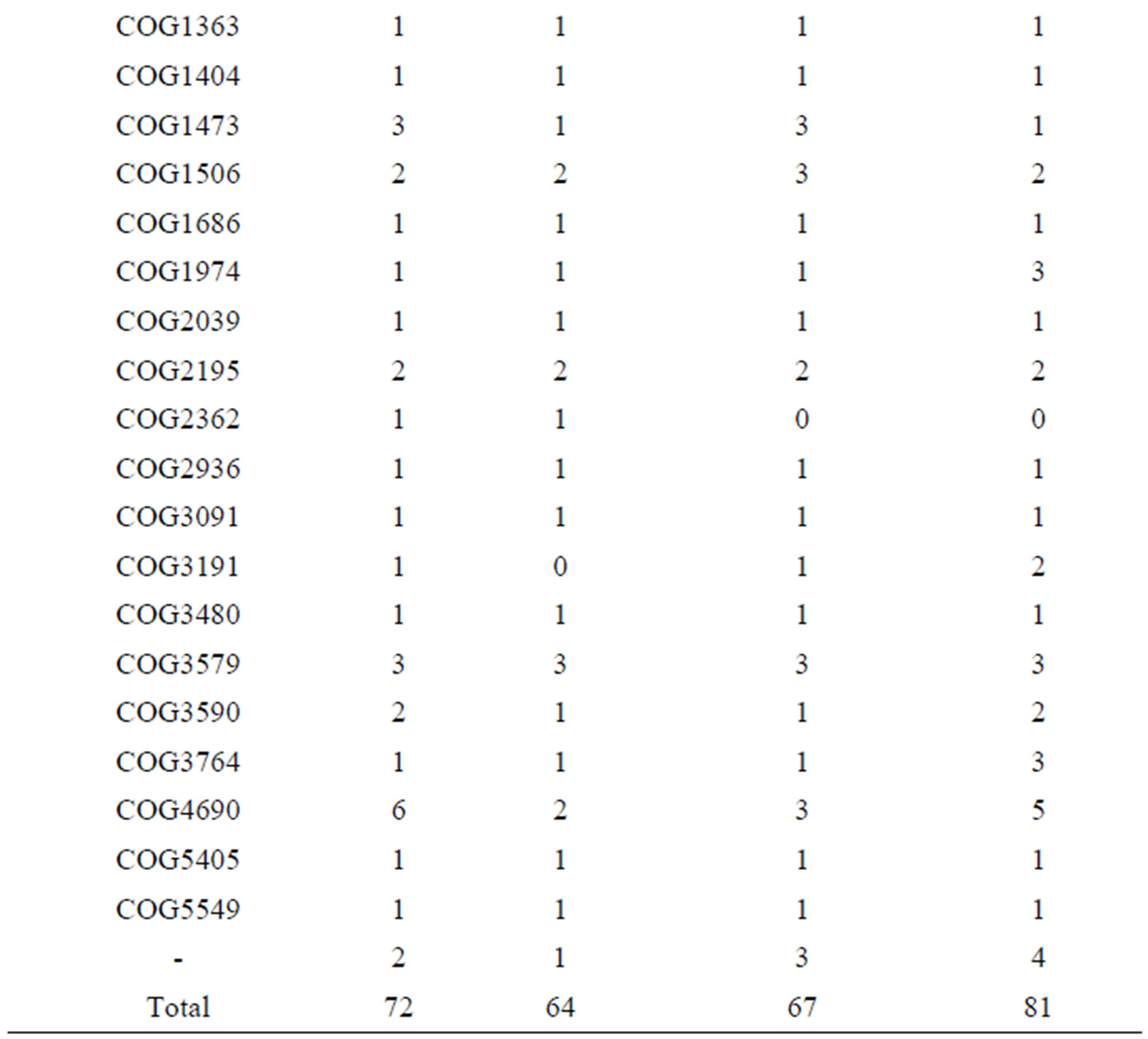
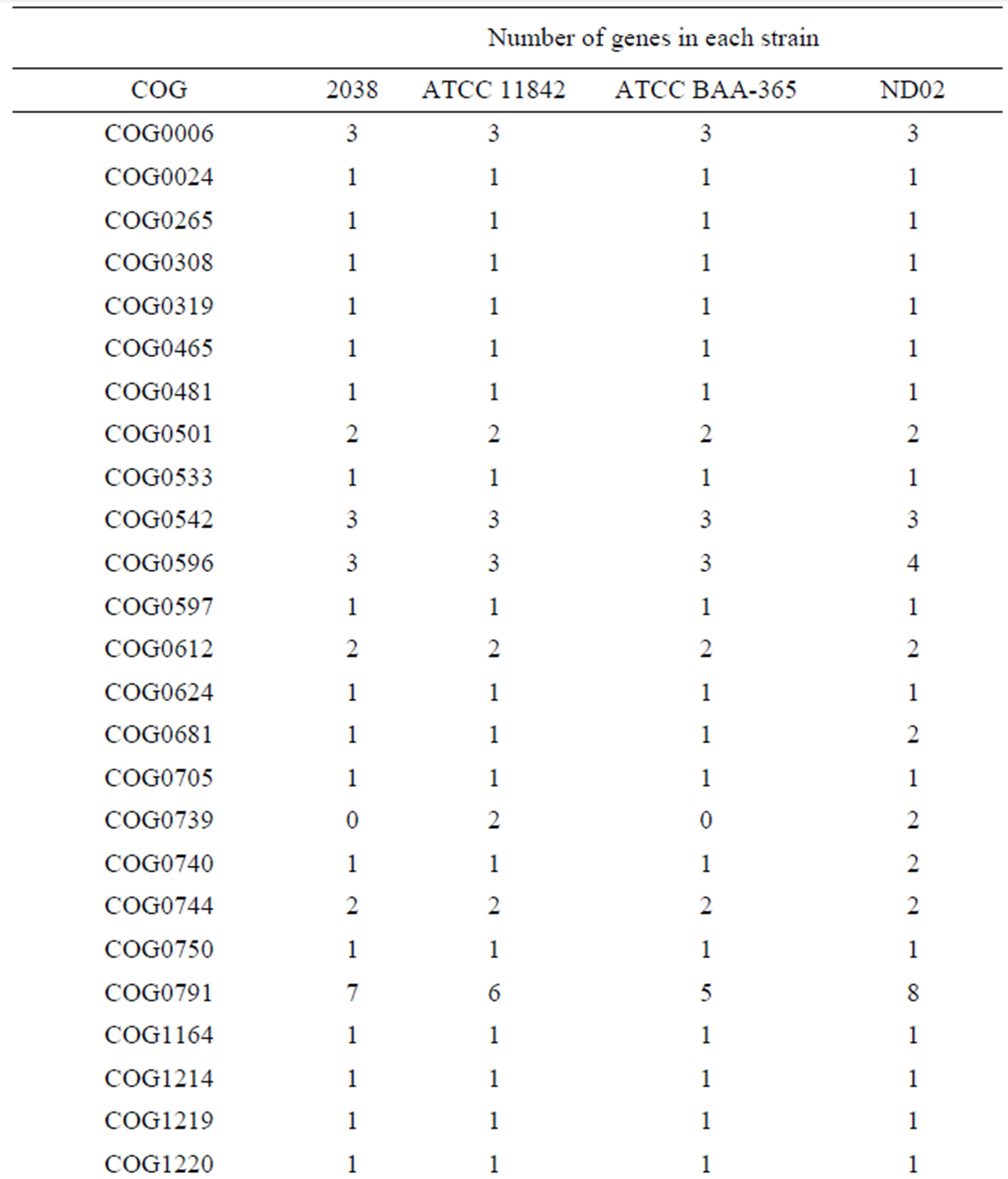
Table S2. Genes encoding proteases and peptidases in each L. bulgaricus strain.
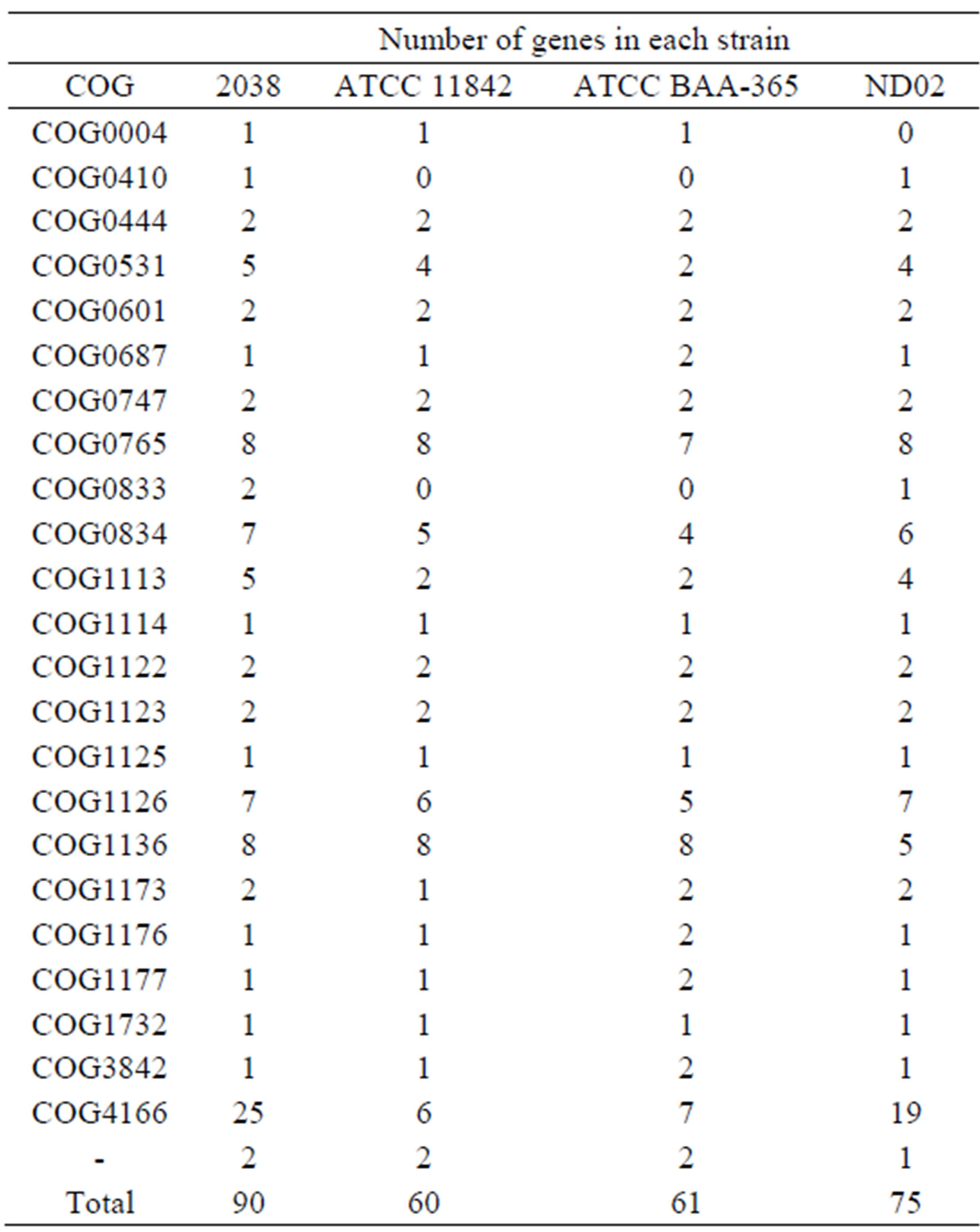
Table S3. Genes encoding peptide or amino acid transporters in each L. bulgaricus strain.
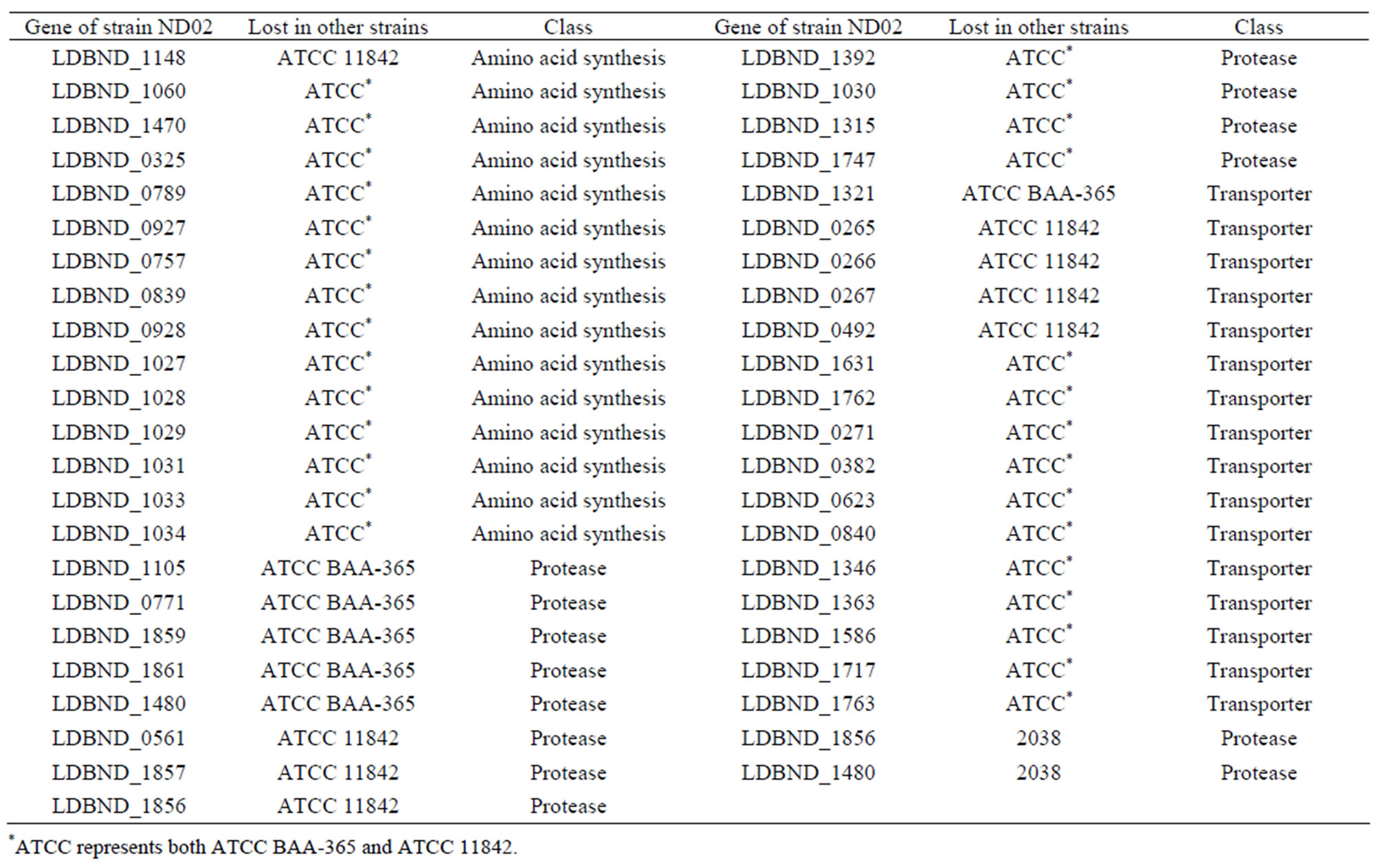
Table S4. Genes involved in proteolysis and amino acid biosynthesis of L. bulgaricus ND02 but lost in other ATCC strains and 2038.
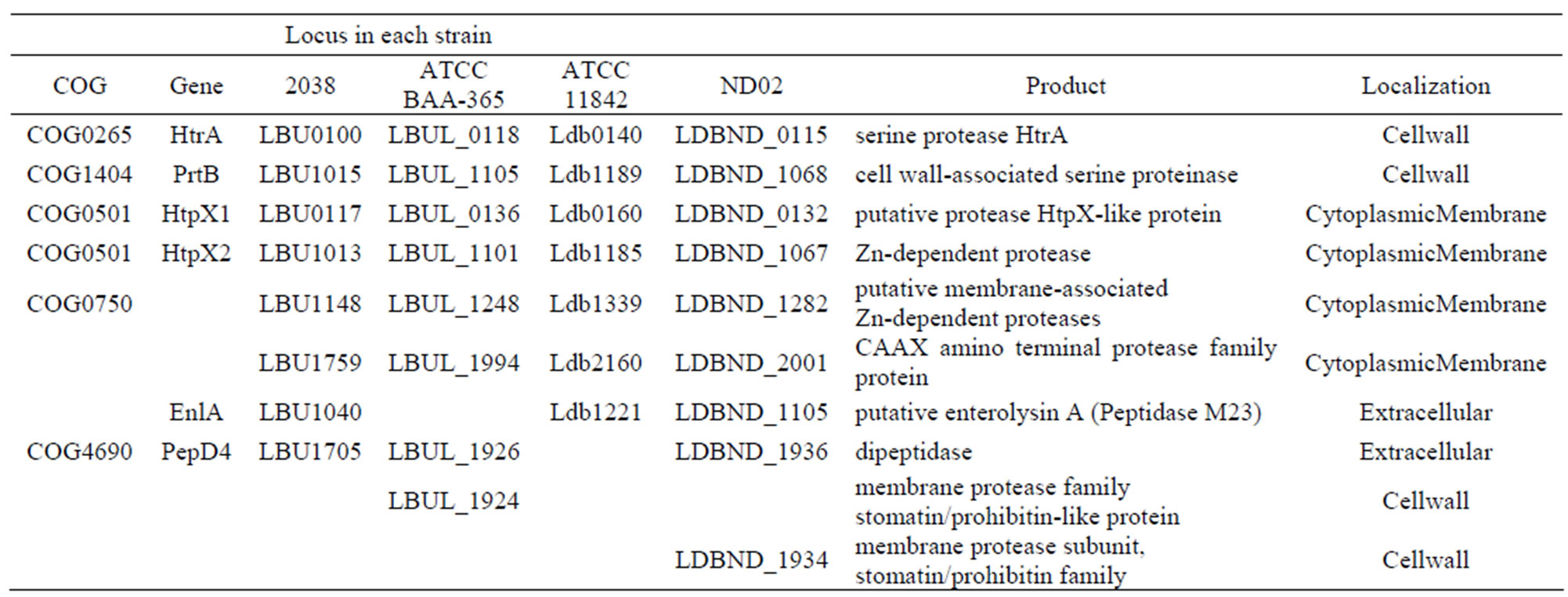
Table S5. Genes encoding extracellular proteases or peptidases in each L. bulgaricus strain.
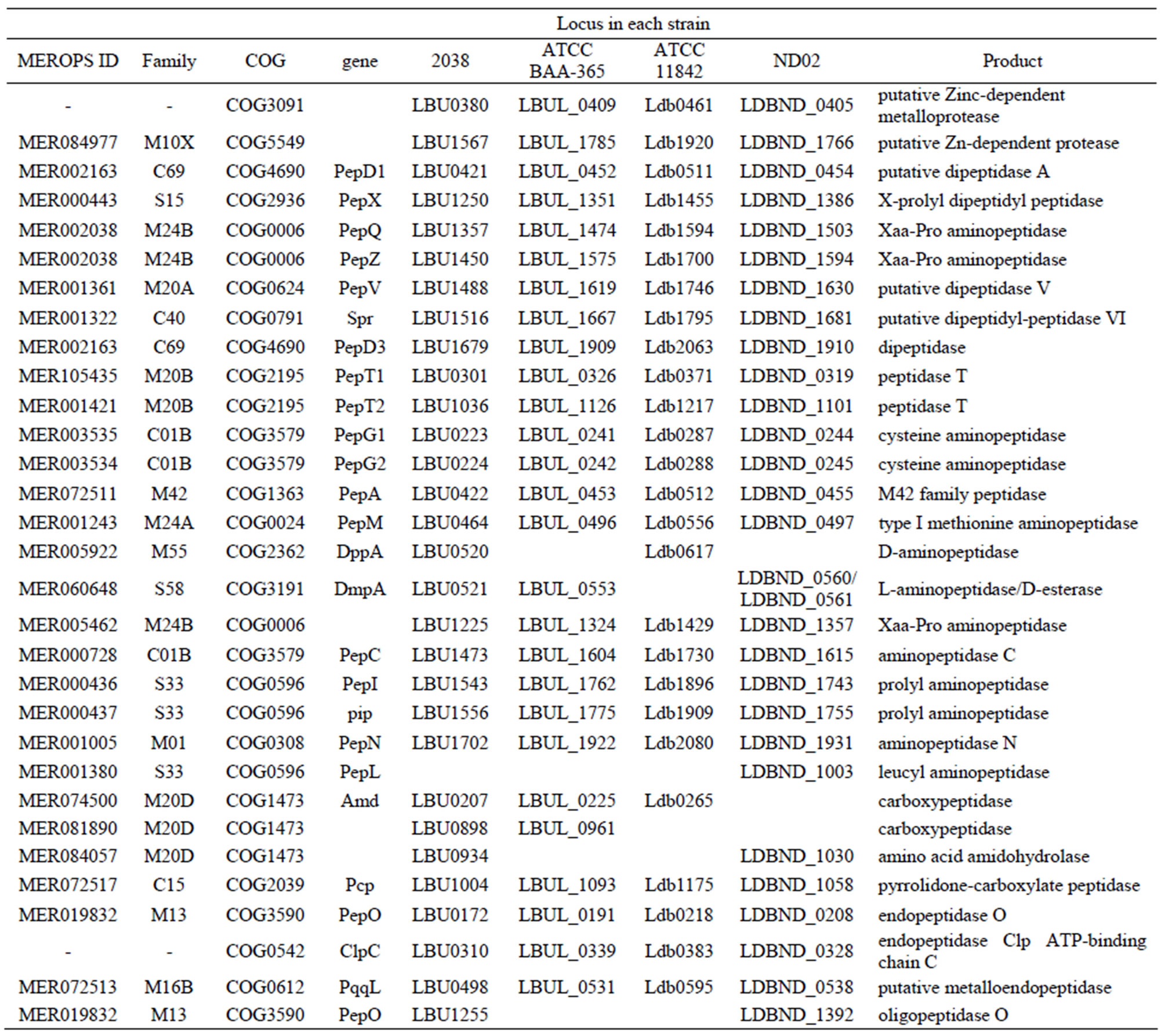
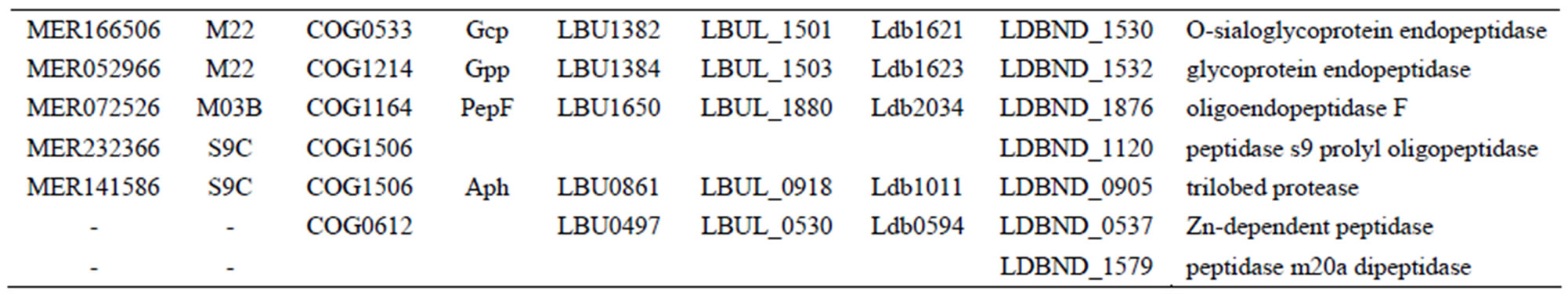
Table S6. Genes encoding intracellular peptidases in each L. bulgaricus strain.
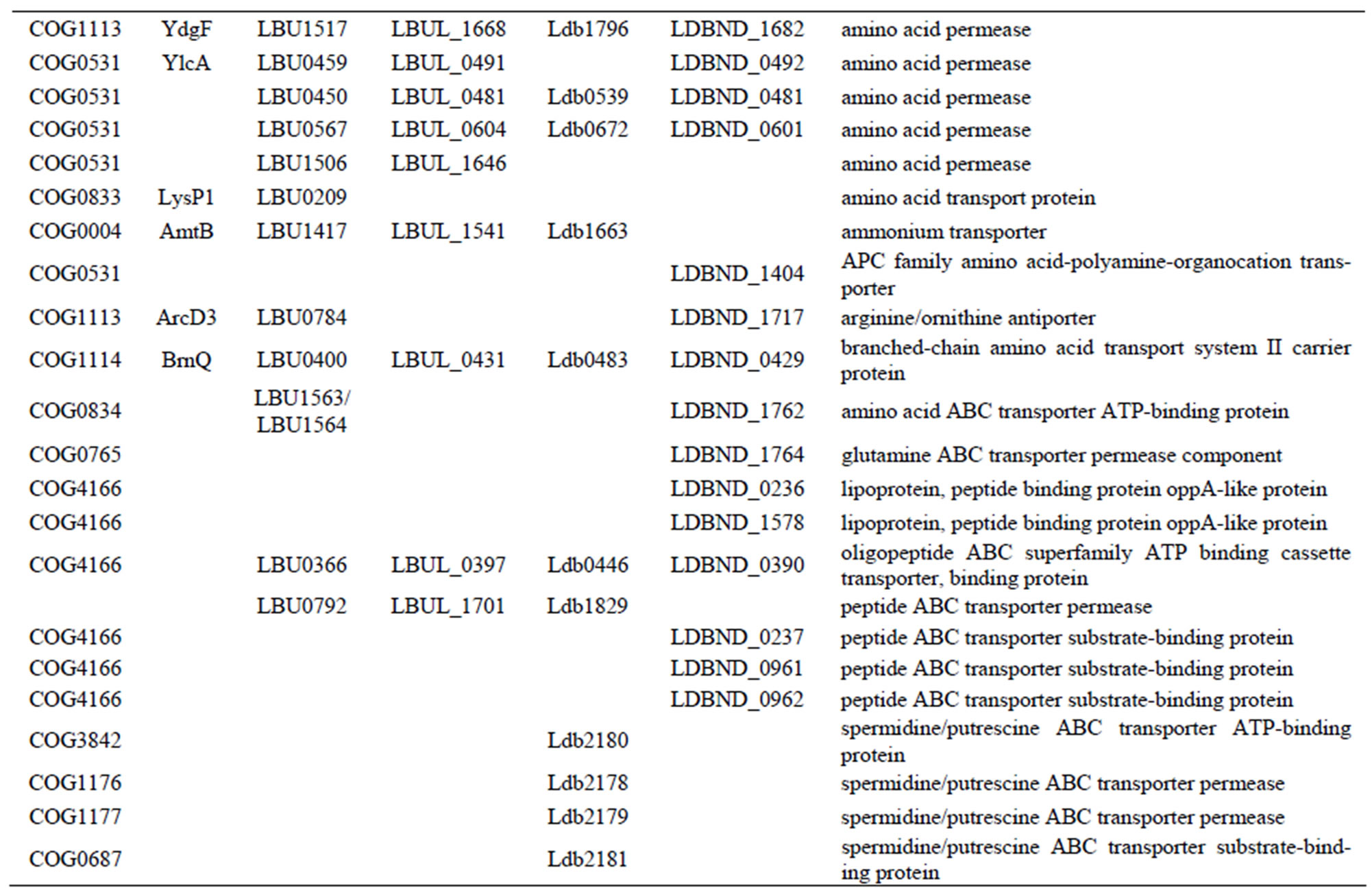
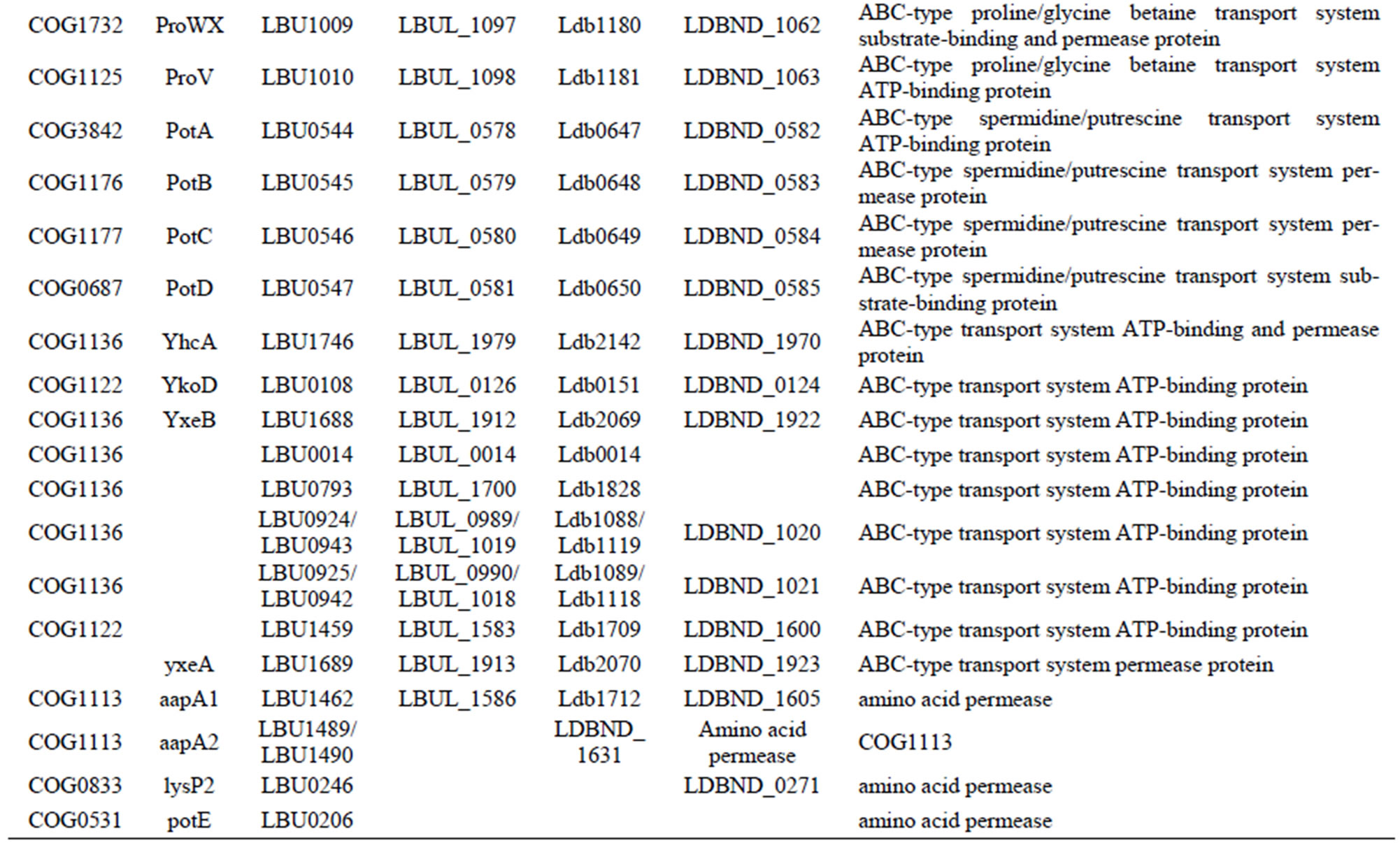
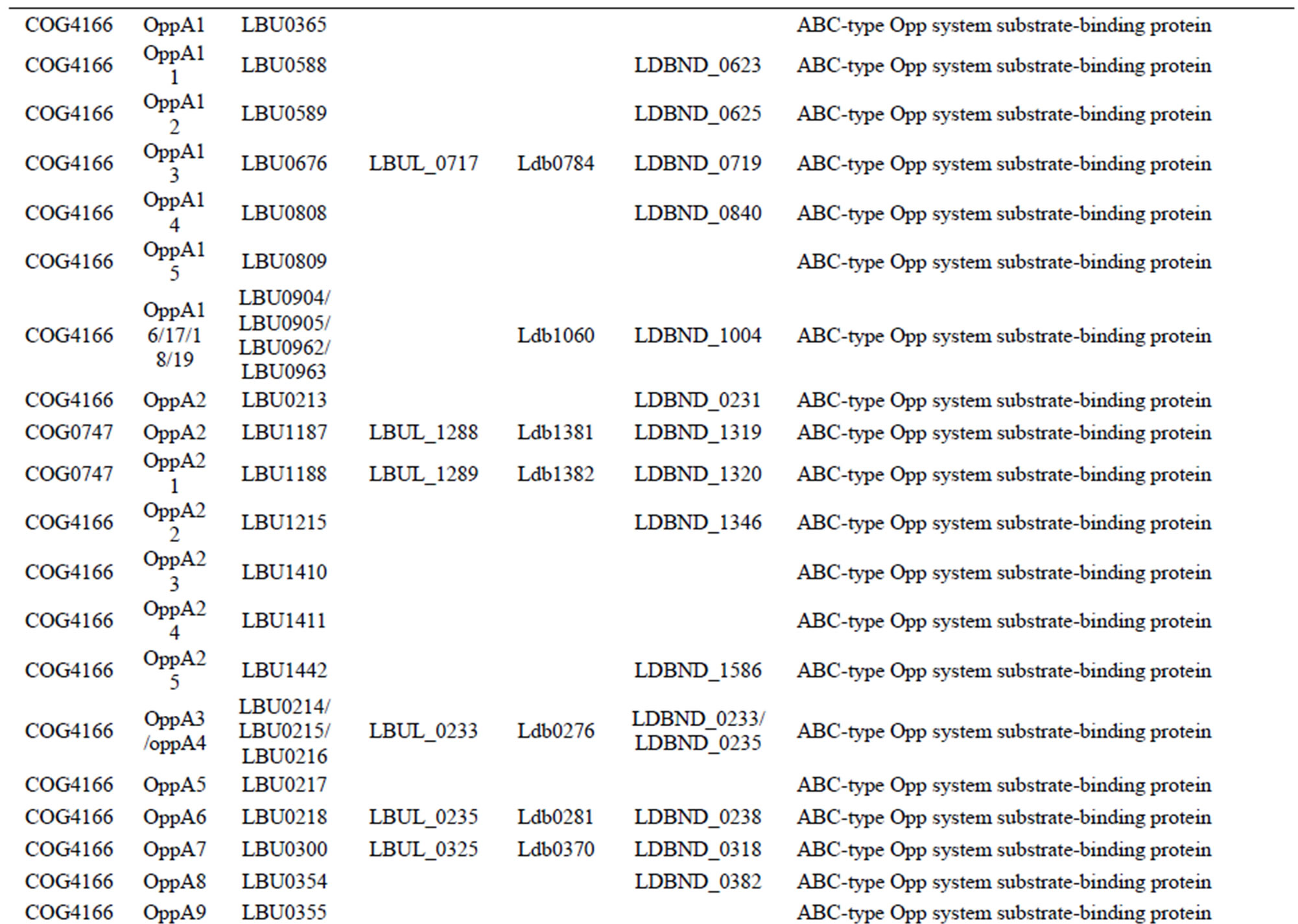

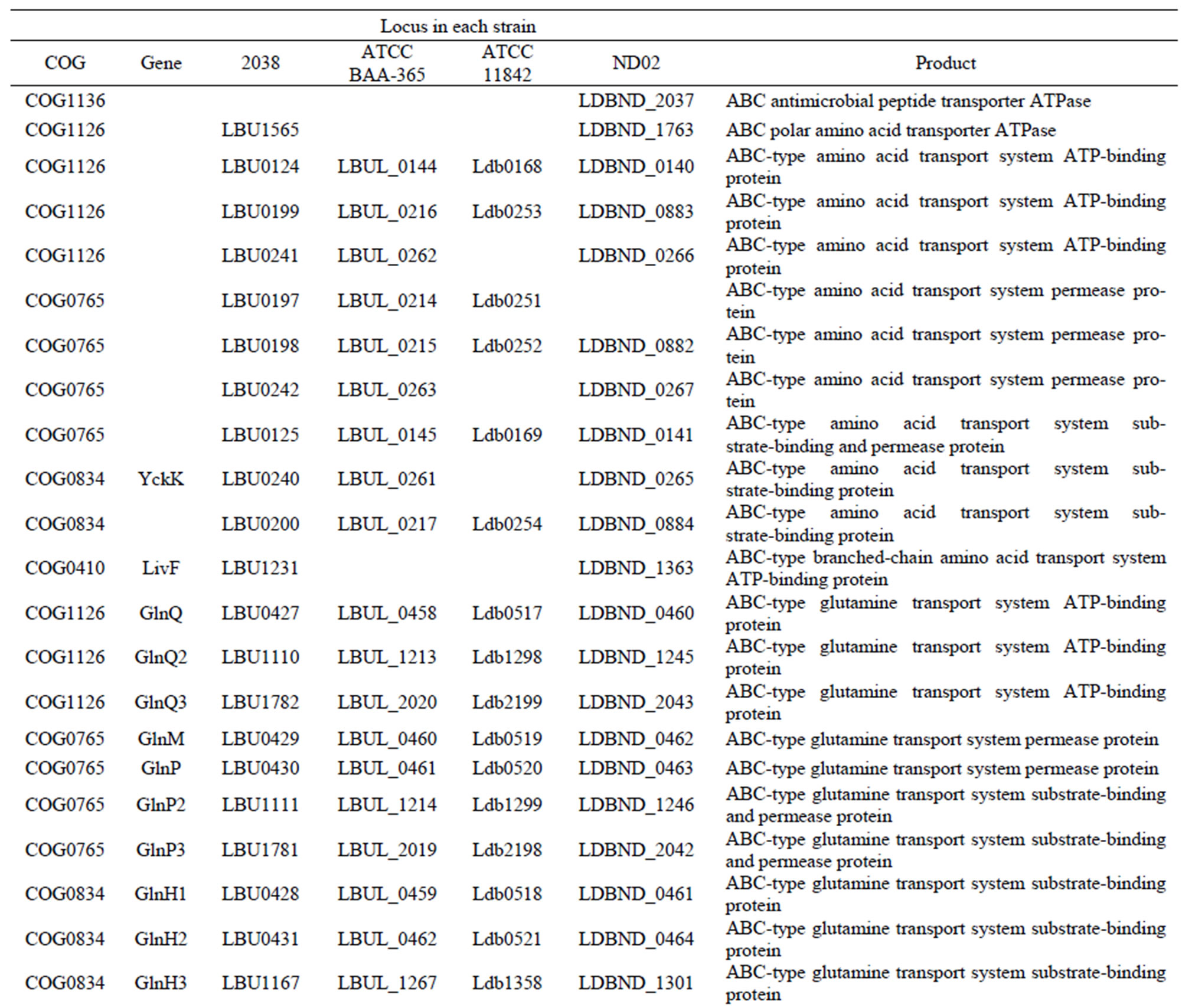
Table S7. Genes involved in oligopeptide or amino acid transport systems in each L. bulgaricus strain.
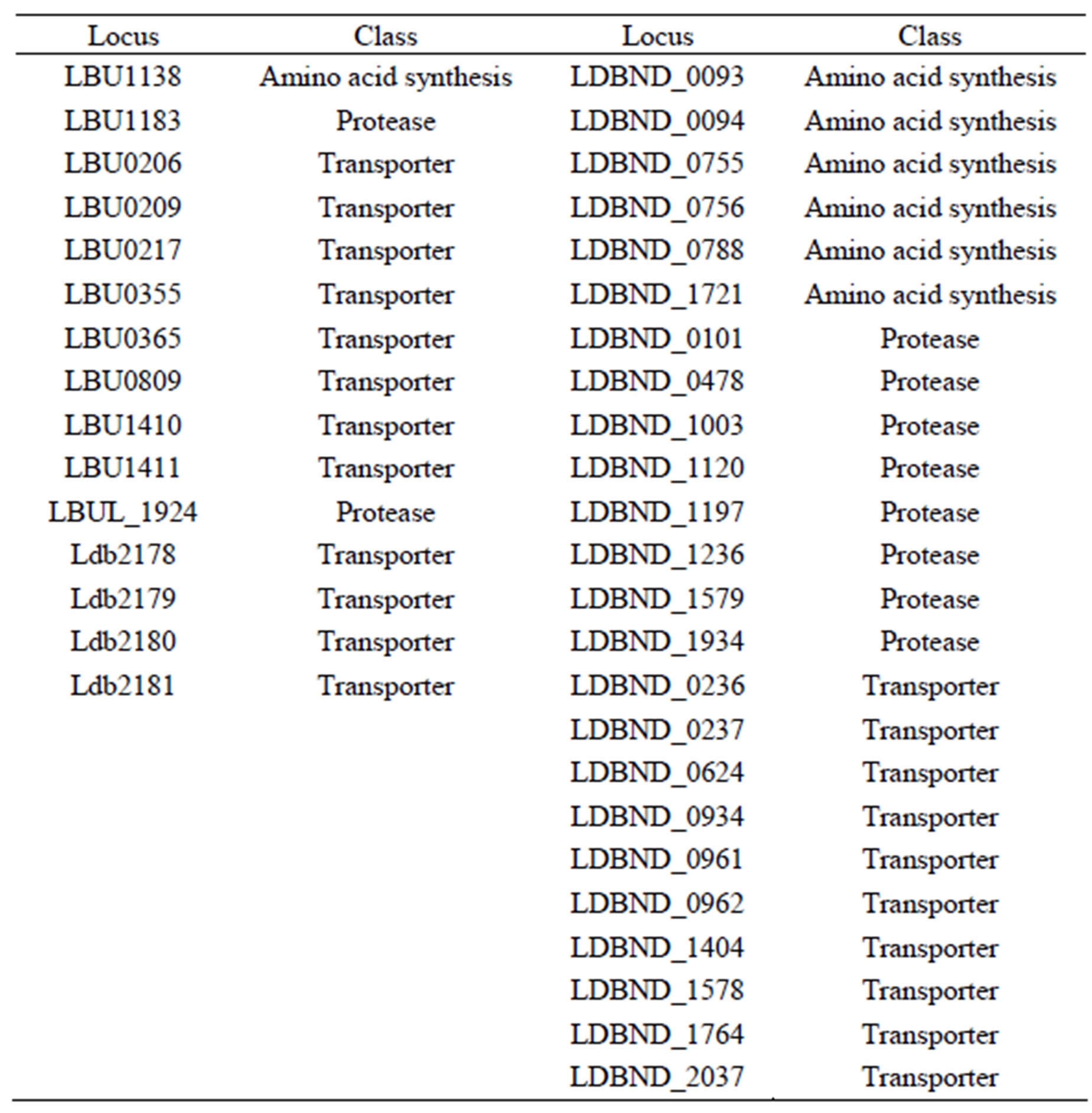
Table S8. Unique genes of each L. bulgaricus strain.
NOTES
*Corresponding author.

