1. Introduction
Many benthic invertebrates can survive in organically-enriched waters, and are often considered as indicators of
pollution [1] . Deposit feeders like nereids live and feed directly on recently deposited sediments, and therefore, are constantly exposed to pollutants [2] . Nereids are the polychetes belonging to phylum Annelida abundant in all environments of the world’s oceans notably successful in mud and sand habitats [3] . The microbial community in the gut of these animals undergoes specific changes to strengthen members that are capable of tolerating and possibly metabolizing toxic compounds encountered by the host gut [4] [5] , and thereby contributing to detoxification of this micro environment. In this process certain groups of bacteria in the gut get selected or rejected depending upon their ability to survive the exposure to pollutants [6] [7] . Li et al. [8] described these remarkable changes in the microbial communities of foregut, midgut and hindgut of Neanthes sp. [7] . Though few attempts have been made to understand these bacterial communities using culture based methods [8] [9] , the relative difficulty in simulating growth conditions of the gut necessitates the use of cultivation-independent methods, in addition to culture based methods. Here we have employed both culture dependent and independent method to understand the bacterial communities in the gut of the Neanthes chilkaensis (Southern, 1921), a polychaete commonly found in the Indian Ocean. The physiological properties of the samples of the nereid isolation were similar to those as described by Vijay et al. [10] and Jaiswar et al. [11] . The culturable gut isolates were checked for their ability to degrade common pollutants which are known to find their way to Gorai mudflat due to idol immersion [12] , from ferries [13] or from the industrial belt located upstream. The results here emphasize the need for studying the micro flora inhabiting various benthic fauna with potential in environmental and industrial applications.
2. Materials and Methods
2.1. Sampling of Animals
Wild Neanthes chilkaensis specimens were collected from the Gorai mudflats (19˚14'0"N; 72˚49'25"E) from a depth of around 5 - 10 cm. The worms were transported in Gamma-sterilized Uricol bottles (HiMedia Laboratories, India) along with native sediment and water. This species could not be found in any other unpolluted sampling locations sampled by us for sediments and water.
2.2. Preparation of Nereid Gut Homogenate
Six mature specimens were randomly chosen from a collected lot of around sixty individuals. Six specimens were placed in a sterile petri dish containing roughly 30 ml filter-sterilized (0.22 μm PVDF membrane, Millipore, France) seawater for 24 hours to allow them to empty their gut contents. While six individuals were placed in a Petri dish containing sterile distilled water for 1 h with water change every 10 min. The worms were starved for 24 h and were then washed in sterile physiological saline (0.85% NaCl), surface sterlized in 2% hypochloric acid [14] , rinsed again with saline and pinned to a sterile cork dissection pad. A fine ventral incision was made about 5 mm from the head continuing up to the tail using a sterile surgical blade, with careful separation of the gut from skin and connective tissue. The worms were rinsed periodically with sterile saline to keep the tissue hydrated. The isolated midgut sections were homogenized in sterile saline using micro-pestles (HiMedia Laboratories). Halves of the midgut homogenates from each of the six specimens were used for total DNA isolation and the other halves were used for culturable studies. Both aspects of the study were conducted simultaneously.
2.3. Isolation of Bacterial Flora from the Midgut Homogenates
One-hundred microlitre each of ten-fold dilutions made in physiological saline, of the six midgut homogenates was spread-plated on Nutrient Agar (HiMedia Laboratories) prepared in sterile aged sea water (adjusted to pH 5, 7 and 8 individually in triplicates) prepared as given before. Plates were incubated aerobically for ten days at 25˚C and the colonies grown were selected on the basis of colony characteristics. Representative colonies isolated from the gut were preserved in glycerol stocks at −80˚C for further analysis.
2.4. DNA Extraction
The isolates obtained from midgut homogenates were grown in tubes containing 5 ml of Nutrient Broth (HiMedia Laboratories), prepared in sea water at 30˚C for 48 hours. The bacterial cells were pelleted by centrifugation (6000 × g for 5 min) and DNA was extracted using the method proposed by Neumann et al. [15] . Community DNA from the midgut of N. chilkaensis the DNEasy Blood and Tissue Kit (Qiagen, Germany) was used with the protocol modified for maximum recovery of DNA from Gram-positive bacteria, as per the manufacturer’s instructions. The quality of the preparations was checked by agarose electrophoresis.
2.5. DGGE Analysis and the Selection of N. chilkaensis Host for Study
Out of the six N. chilkaensis hosts selected, only one host was used for thorough investigation. This was done by first comparing the Denaturing Gradient Gel Electrophoresis (DGGE) profiles of 16S rDNA amplicon of the mid-gut bacterial community of six hosts and then selecting the consensus one. Touchdown PCR was used to amplify the eubacterial hypervariable region V3, of the 16S rRNA gene using 338F (5'-ACTCCTACGGGAG- GCAGCAG-3') and 518-GC (5-ATTACCGCGGCTGCTGG CGCCCGCCGCGCGCGGCGGGCGGGGCGG- GGGCACGGGGGG-3') [16] . The PCR reaction mixtures (50 μl) contained 50 - 100 ng of template DNA, 200 µM of each dNTP, 10 mM of each primer, 10% of 10× Taq Buffer, 25 mM MgCl2, 1.5 units of Taq DNA polymerase (Bangalore GeNei, India) and sterile deionized water (Milli-Q). The touchdown PCR conditions were 94˚C for 2 min; 20 cycles of 94˚C for 30 sec followed by a 62˚C - 57˚C touchdown for 30 sec and 72˚C for 1 min. This was followed by 10 cycles of the same temperature steps with the touchdown replaced with a constant annealing temperature of 57˚C for 30 sec. The above program was followed by a final extension for 15 min at 72˚C. DGGE gels were run on the DCode electrophoresis unit (BIO-RAD, Singapore). For each sample, 400 ng of the PCR product was loaded on a 1-mm thick, 10% polyacrylamide gel containing 40% to 70% gradient of denaturant (urea and formamide). Electrophoresis was carried out for 18 hrs at 70 V, in 1× TAE buffer, at a constant temperature of 60˚C. The gels were silver-stained and visualized in white light (DNR Bio-Imaging Systems, Israel). DGGE fingerprints obtained from the six individuals were compared using the Renkonen index [17] of similarity, a percentage based measure of overlap between communities, using the Community Analysis Package 4.0 (Pisces conservation Ltd., UK), which is defined as
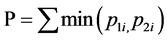
where, P is the similarity between community 1 and 2. p1i is the proportion of group i in 1; p2i is the proportion of group i in 2.
The individual bands representing distinct clones were carefully dissected from the gel, purified using QIAquick Gel Extraction Kit (Qiagen) and the elutes were preserved at −20˚C for further analysis. One of the samples, selected to represent the common DGGE banding pattern, was continued with cultivation-independent analyses, and its isolates were taken for cultivation-dependent studies.
2.6. PCR Amplification of the 16S rRNA Genes
Eubacterial primer set 63f (5'-CAG GCC TAA CAC ATG CAA GTC-3') and 1387r (5'-GGG CGG WGT GTA CAA GGC-3') [18] were used to amplify 16S rRNA genes from isolates and midgut total DNA with a QB96 Thermocycler (Quanta Biotech, UK). The thermal cycling conditions followed were initial denaturation at 94˚C for 4 min; 30 cycles of 94˚C for 30 s, 57˚C for 30 sec and 72˚C for 1 min 30 sec and a final extension step of 72˚C for 30 min. The amplified products were eluted using the QIAquick Gel Extraction Kit (Qiagen). The eluates were cloned using the same method described below, sent for sequencing to Bangalore GeNei, Bengaluru, India, and sequences were checked against the GenBank database using the NCBI-BLAST program.
2.7. Construction of the 16S rRNA Gene Clone Library
The eluted ~1.3 kb amplicons obtained from the midgut community DNA, were cloned (in duplicate) into the pTZ57R/T vector (Fermentas, USA) according to the manufacturer’s instruction, and transformed into Escherichia coli DH5α, using the InsTA-cloning kit (Fermentas). The presence of the inserts in the colonies was checked by PCR using M13 standard sequencing primers (Fermentas). The amplicons obtained were digested using HaeIII and HpaII (Fermentas) and screened by Amplified rDNA Restriction Analysis (ARDRA) in order to avoid sequencing of redundant clones. Multiple representatives of each ARDRA group were selected and sequenced commercially (Bangalore GeNei).
2.8. Phylogenetic Analysis of the 16S rRNA Gene Clone Library
Single parse sequence analysis with approximately 700 - 850 base reads were corrected using the Chimera Check program at the RDP website (http://rdp.cme.msu.edu/). The partial 16S rRNA gene sequence reads from the cultivation-dependent and independent studies were compared against the submissions available in GenBank (NCBI), using the BLAST algorithm [19] . Phylogenetic analyses were conducted using the MEGA4 Package [20] and the ClustalW interface of MEGA4 was used to make multiple alignments. The evolutionary history was inferred using the Neighbour-Joining method [21] . Homologous regions of 560 bp (position 93 to 653), corresponding to the V2-V3 region in E. coli ATCC 11775T were used for phylogenetic analysis based on the understanding that phylogenetic trees constructed using partial sequences have comparable topologies with those constructed using full sequences [22] [23] . From phylogenetic tree, operational taxonomic units (OTUs) were studied.
2.9. Abilities of the Isolates to Utilize Recalcitrant Aromatic Compounds as a Sole Carbon Source
The isolates were checked for their ability to utilize the following polyaromatic hydrocarbons (PAHs) like benz(k)fluoranthene (B(k)F), benz(k)pyrene (B(k)P), chrysene (CHY), dibenz(a,h)anthracene (DBA), fluorene (FLU), naphthalene (NAF), phenanthrene (PHE) and pyrene (PYR). The PAHs were supplemented in mineral media as sole carbon sources. 250 μl of each of the filter sterilized 0.4% polyaromatic hydrocarbons made in dimethylformamide (DMF) were spread separately on the surface of MM1 agar (containing (L−1), 2.92 g KH2PO4; 1.51 g K2HPO4; 1.0 g (NH4)2SO4; 2.02 g CaCl2∙2H2O; 2.26 g Na2HPO4∙7H2O; 20 μg FeCl3∙6H2O; 3.12g MgSO4∙7H2O and 20 g of agar) plates [24] . To rule out the DMF degradation by the bacteria, the isolates were also streaked on control plates with 250 μl DMF spread on it. 2,4,6-trichlorophenol (TCP) utilization was checked using basal liquid medium (MM2) (containing (L−1) 0.5 g H3BO3; 0.4 g ZnSO4∙7H2O; 0.4 g MnSO4∙H2O; 0.2 g FeCI3 6H2O; 0.2 g (NH4) 2MoO4; 0.1 g KI; and 0.04 g CuSO4∙5H2O) [25] . 250 μl of filter-sterilized 1% stock of TCP made in 0.2 N NaOH, was added aseptically to 20 ml of autoclaved MM2 to serve as the sole carbon source. About 108 cells of each of the isolates were inoculated in this medium and were incubated at 25˚C for three weeks. Turbidity suggested the growth of the organism. The inference was confirmed by observing growth on MM2 agar plates along with TCP. Isolates capable of utilizing TCP as the sole carbon source could also grow without the biotin supplement.
2.10. Assay for Surfactant Tolerance and Production
Isolated bacteria were assayed for surfactant resistance according to Plante et al. [9] using CTAB and Triton-X100 just above the general toxic level for microorganisms. Hemolysis was used as an initial screening criterion for biosurfactant-production by the isolates. The isolates were streaked on Blood Agar (HiMedia Laboratories) and incubated at 30˚C for 48 hours after which they were checked for hemolysis. Since hemolysis is only a suggestive method for studying biosurfactant production by bacteria [26] , surface tension was measured for each isolate using a surface tensiometer (Khushboo Scientific, India) employing the DuNouy’s platinum-ring method. The bacterial isolates were grown in 250-ml Erlenmeyer flasks, in triplicates, containing 50 ml Luria- Bertani broth (HiMedia Laboratories) at 30˚C on a rotary shaker (180 rpm) for 24 hours. The cultures were centrifuged (8000 × g) for 10 min and cell-free supernatants were checked for the reduction in the surface tension at room temperature. Emulsification index (% EI) of the supernatants was calculated by the method described by Markande et al. [27] using the formula
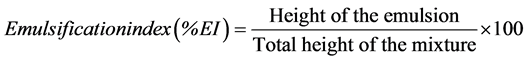
3. Results
3.1. Bacterial Communities in the Mid Gut of N. chilkaensis Hosts
The DGGE banding patterns (Figure 1) obtained for the gut bacterial community of the six N. chilkaensis individuals were highly similar indicating a regular bacterial community being native to the specimens. The similarities are further evident from the Renkonen indices (Table 1). The sample No 2 was selected as a representative sample for furthering detailed analysis.
![]()
Figure 1. Denaturing gradient Gel Electrophoresis (DGGE) profiles of V3 region of six different N. chilkaensis midgut samples. Lanes are labeled with midgut sample numbers.
![]()
Table 1. Renkonen indices for midgut bacterial community similarity between different N. chilkaensis hosts.
aWm1, Wm2, Wm3, Wm4, Wm5, Wm6 represent midguts isolated from six N. chilkaensis individuals.
The nutrient agar plates having different pH were carefully observed for colonies. All the isolates picked up could be grouped into 13 colony morphotypes. All these representative morphotypes were identified by sequencing of 16S rRNA gene and BLAST analysis. The isolates were largely firmicutes, the majority of which fell within the genus Bacillus (Table 2). The sequences from few of the isolates matched at varying percentages with a PAH-degrading Bacillus sp. (98.4% to 99.8%). While sequence from isolate GNG-AV15 matched to that of a Staphylococcus sp., GNG-AV13 matched a Kytococcus sequence (Table 2).
3.2. Phylogenetic Analysis of 16S rRNA Clone Library
A total of 108 clones were found to be positive for the 1.3-kb insert. The patterns obtained from ARDRA were used to construct a dendrogram wherein the 108 clones were grouped into eight large ARDRA clusters. From each group, a minimum of two and a maximum of eight clones were selected for sequencing. ARDRA branches represented by single clones were also sequenced. A total of 49 clones were analyzed by Single Parse Analysis, of which two were revealed as possible chimeras. This left 47 representatives of the original 108 clones. The clone coverage could, thus, be calculated by Good’s formula [28] for the 12 taxa (Table 3 and Figure 2) among the original 108 clones. The coverage was found to be 90 percent, which means that there was a 10 percent chance that the 109th clone would yield a new OTU. Four clones in the original library were present as single copies and contributed to some loss of coverage.
The diversity observed among isolates and clones spans across many phyla, including Proteobacteria, Firmicutes and Actinobacteria. The 16S rRNA gene sequences from the isolates and clones were aligned with their
![]()
Table 2. Phylogenetic affiliation of isolates frome midgut of N. chilkaensis.
![]()
Figure 2. Phylogenetic affiliations of the members of the bacterial community residing in the midgut of N. chilkaensis.
![]()
Table 3. Phylogenetic affiliation of clones recovered from the midgut of N. chilkaensis.
nearest phylogenetic relatives from GenBank and a dendrogram was constructed (Figure 2). Identical OTUs in the cultural studies were also detected in the culture-independent study. GNG-SV21, GNG-SV91, GNG-SV112 were found to belong to the same taxon of Bacillus sp. (FJ449840) as isolates GNG-AV4, GNG-AV6, GNG- AV7, GNG-AV9, and GNG-AV12 (Table 2 and Table 3) covering 20.4 percent of the library. About 12 percent of the clone library matched with a Phyllobacterium sp. (AM989040), 7.4 percent with an Agrobacterium sp. (FJ425916), 11.1 percent with a DEHP-degrading Gordonia sp. (DQ301916), 10.1 percent with napthalene utilizing Microbacterium laeveniformis sp. (AF535159) and 12 percent with an organo-chromium complex degrading Leifsonia sp. (DQ901014). About 3.7 percent of the library (Table 3) matched uncultured entries on Genbank with 92.8% and 97.1% similarities. Having observed the presence of bacteria and clones matching with isolates capable of degrading recalcitrant hydrocarbons, it was decided to study the abilities of the isolates to degrade common aquatic pollutants.
3.3. Recalcitrant Hydrocarbon Degrading Abilities of the Isolates
Bacteria that are able to degrade hydrocarbons may use them as carbon source. Thirteen shortlisted isolates were tested for their abilities to grow with benz(k) fluoranthene (B(k)F), benz(k) pyrene (B(k)P), chrysene (CHY), dibenz (a,h) anthracene (DBA), fluorene (FLU), naphthalene (NAF), phenanthrene (PHE), pyrene (PYR) and 2,4,6-trichlorophenol (TCP), as sole carbon sources on mineral media agar plates and their growth is given in Table 4. Bacillus sp. GNG-AV7 and bacterium GNG-AV14 were unable to utilize any of these complex substrates. Isolate Staphylococcus sp. GNG-AV15 was able to utilize most of the PAHs, but failed to use TCP. Bacillus sp. GNG-AV4 and Bacillus sp. GNG-AV6 were also able to utilize majority of the PAHs. The only isolate that could utilize almost all the hydrocarbons tested including TCP, was GNV-AV8, belonging to Bacillus sp. (Table 4).
3.4. Resistance and Production of Biosurfactant
Although most of the bacteria isolated showed resistance to CTAB, only Kytococcus sp. GNG-AV13 and Staphylococcus sp. GNG-AV15 were found to be resistant to Triton-X100 (Table 5). It is recognized that the emulsification of the nonpolar hydrocarbon with water is the first step that helps bacteria to make hydrocarbons available as carbon source. Many of our isolates were found to be capable of producing biosurfactants. The amount of biosurfactant produced by each isolate was quantified after growth in Luria-Bertani broth for 24 hours. Only Bacillus sp. GNG-AV8 was found to reduce surface tension substantially, to 31.7 ± 1.2 mN/m from an initial value of 60 mN/m for sterile medium (Table 6), while Bacillus species GNG-AV1, GNG-AV7, GNG- AV11, G-AV12 and Staphylococcus sp. GNG-AV15 could be considered as mediocre biosurfactant-producers as compared to the former (Table 6).
4. Discussion
Recent surveys on polychaetes in the region of Mumbai [29] - [31] do not record the presence of Neanthes chilkaensis. The species of Nereids recorded from this region so far include Perinereis, Platynereis and Dendronereis [31] . The presence of N. chilkaensis to the tune of 85 percent of the Nereid population in Gorai was surprising. The species was not observed in any of our samplings from other areas of Mumbai waters. The present study was undertaken to understand the nature of the gut bacterial communities and to investigate their roles in alleviation of pollution stress of these hosts.
Keeping in mind the difficulties to culture some bacteria outside their native environment [28] - [30] , we employed both cultivation-dependent and cultivation-independent molecular approaches. Cultivation dependent and independent findings have often been disparate with respect to Firmicutes and Actinobacteria [32] - [35] and the reasons attributed to such discrepancies are many. König [36] noted that Gram positive bacteria take part in early and intermediate steps of degradation. In this, the sporulating members may grow on plates but their inefficient lysis during DNA extraction may exclude them from being detected in the cultivation-independent study. On the other hand, more rigorous protocols for recovery of Gram-positive DNA could be successful at the cost of damage to Gram-negative DNA [37] . Nevertheless, these factors may lead to a biased under-representation of Gram-positive bacteria in clone libraries, especially felt in niches where their presence could be relevant.
Among the six individual hosts selected, a clear similarity was evident from the DGGE patterns (Figure 1). It is therefore likely that the community profile represents microbiota that survived in harsh gut microenvironment,
![]()
Table 4. Ability of the isolates to degrade different Poly aromatic hydrocarbons (PAH).
Benz(k)pyrene or B(k)P, dibenz(a,h)anthracene (DBA), Phenanthrene (PHE), Fluorene (FLU), Chrysene (CHY), Naphthalene (NAF), Benz(k)fluo- ranthene (B(k)F); and 2,4,6-Trichlorophenol (TCP). PAH degraders (+) and non-degraders (−) are denoted.
![]()
Table 5. Surfactant resistance by isolates.
Growth (+): resistant; no-growth (−): sensitive.
suggesting that the residing (or attached) communities may not be a random mix of opportunists from each host’s immediate environment. This lack of variability also provided the rationale for proceeding with a representative individual. Moreover, the 16S rRNA clone library generated can be considered to be representative of the residing bacterial assemblage in the midguts of the pollution-enduring N. chilkaensis populations of Gorai.
In this study, 53.7 percent of the clones match Gram-positive entries of GenBank (Table 3). At the same time all the isolates belong to members of Gram-positive groups i.e. Actinobacteria and Firmicutes, indicating a dominance of firmicutes in the midgut of N. chilkaensis. It was interesting to find about 20.4 percent of the clone library and 40% of the isolates matched with a PAH-degrading Bacillus isolate (FJ449840) (Table 3).
![]()
Table 6. Surface tension (ST) and emulsification index (%EI) of the culture supernatants of the midgut isolates, grown under identical conditions.
aThe surface tension of the uninoculated medium was 63 ± 0.4 mN/m, with freshly inoculated broth having values within the same range.
Few isolates (Table 2), did not represent the clone library were Staphylococcus isolate (GNG-AV15) and Kytococcus isolate (GNG-AV13). As expected many of the isolates are reported as cultured and uncultured members from other marine invertebrates (Table 2).
The GenBank relatives of Leifsonia, Microbacterium and Gordonia detected by the cultivation-independent approach are reported to have unique degradative abilities (Table 3). None of them, however, were present among the cultured isolates (Table 2). Interestingly, these three entries also happen to be close phylogenetic relatives of three PAH-utilizing isolates from a previous study [38] . It is probable that we have missed some bacteria belonging to Actinobacteria phylum in the culturable study because the selective procedures such as heat- treatment and incorporation of specific substrates necessary for isolation of actinomycetes, were not used. In similar studies of Nereid midgut phylogeny Li et al. [8] observed only two Gram positive bacteria (Clostridium species).
While Gram-negative matches constituted about 43 percent of the clone library, none of the midgut isolates was Gram negative. One of the two other Gram-negative clusters in the clone library matches a dibenzothiophene-degrading Agrobacterium tumefaciens and a 2,4-dichlorophenoxyacetic acid degrading Cupriavidus strain representing 7.4% and 11.1% of the clonal library respectively (Table 3). The slow growth of these α-proteo- bacterial members might have resulted in their absence on plate as indicated by previous workers [32] [39] .
The Gorai mudflats have become deeply affected by the dumping of wastes directly into the sea, pushing the system to the brink of crisis [13] . Since the polychaete N. chilkaensis were collected from polluted mudflats of Gorai, Mumbai, we have tried to characterize the culturable flora by their abilities to utilize common polyaromatic hydrocarbon pollutants like dibenz(a,h)anthracene, benz(k)pyrene, benz(k)fluoranthene, chrysene, fluorene, naphthalene, and phenanthrene. These chemicals are known to find their way to Gorai mudflat due to idol immersion [12] , from ferries [13] or may come from the industrial belt located upstream. We also tested the abilities of the isolates to utilize 2,4,6-trichlorophenol alongside a range of PAHs because Trichlorophenols such as 2,4,6-trichlorophenol, is widely used by pesticide, glue and the textile industries and had a strong chance of being present in Gorai originating from the small and medium industries located along Gorai creek. Of all the isolates checked for their ability to utilize 2,4,6-trichlorophenol, only Bacillus sp. GNG-AV8 was found to be positive (Table 4).
Many of the midgut isolates (84%) were able to utilize PAHs as sole carbon sources (Table 4). The factors governing PAH utilization by degradative organisms range from the efficiency of uptake mechanisms to the ability to enhance the availability of the compound by biosurfactant and bioemulsifier production [40] - [42] . Nereid guts are shown to have high concentrations of surfactants with numerous chemical structures [9] . While bacteria are known to produce biosurfactants, it is still not clear whether the source of these surfactants is the gut microbial community [43] . As was seen in these results, most of the isolates were resistant to cationic surfactant CTAB, an antiseptic Cetrimide while only few were resistant to non-ionic surfactant Triton-X100 (Table 5). Klosterhaus & Baker [44] studied the role of surfactant micelles in the absorption of chlorinated hydrocarbons and polyaromatic hydrocarbons in polychaete gut. Their study indicates that absorption requires presence of surfactant micelles to solubilize hydrocarbons in gut fluid. About 77 percent of the isolates in the present study were found to considerably reduce the surface tension of Luria Bertani Broth (to or below 55 mN/m), with identical conditions of growth (Table 6) when checked with cell-free extracts. In comparison to Bacillus sp. GNG-AV8 (Table 6), which was able to reduce the surface tension of LB broth by about 49.68 percent, the other isolates can be regarded as mediocre biosurfactant producers with the ability to reduce surface tension in the range of 12.22 to 24.92 percent. From this study it is clear that these isolates could not only utilize the PAHs as carbon source but also produced biosurfactant to make these hydrophobic substances available for degradation. The GNV-AV8 that exceeded other isolates in its biodegradation capabilities was also a better biosurfactant producer. This indicates that degradation is probably related to biosurfactant producing ability. The PAH utilizing bacteria might depend on other bacteria for biosurfactant, but it enhances its independence if they do both the activities. Klosterhaus & Baker [44] reported the likely requirement of micelles in solubilization of sediment bound hydrophobic organic compounds (HOCs) before accumulation in Nereis virens gut having high amounts of surfactants in their gut fluids. Although the cultivation-independent detection of 16S rRNA sequences of various degradative strains from other studies does not necessarily prove their functional relevance, it is interesting to note that a majority of the clones (72.1 percent) detected in the gut of N. chilkaensis, match closely with GenBank entries of many aromatic pollutant degraders or organisms isolated from contaminated sites (Table 3). Evidence from the cultural studies supports the ability of the midgut isolates to degrade a complex array of aromatic recalcitrants with the help of their abilities to produce surfactants. The nature of the bacterial members, specifically the predominance of degraders in the gut community, indicates possible selection of microbial community in the gut of N. chilkaensis with respect to pollution.
Although these benthic organisms have been studied as pollution indicators, their gut flora can yield interesting microorganisms with immense application potential.
The Circle branch tree is further shown in three separate clusters of branches. The sequences were first aligned using the CLUSTALW program and a neighbour-joining tree was generated using the Kimura-2 parameter method for computing the evolutionary distances. E. coli positions 93 - 653 were considered for the analysis. The numbers at the nodes indicate the bootstrap values based on 1000 replicates. The small bar indicates the Jukes-Cantor evolutionary distance. The GNG-AV series represent cultured isolates and the GNG-SV series represent the clones from the culture-independent study. The rest of the members are closest-neighbour matches as recovered from BLAST searches on the GenBank database.
Acknowledgements
The authors gratefully acknowledge the help rendered by Dr. B. Suresh at the Maharaja Sayajirao University (MSU) of Baroda for identifying the specimens used in our study; Dr. A. S. Nerurkar of MSU for the use of laboratory facilities; and Mr. A. K. Padmanabhan and Mr. J. M. Koli of CIFE for technical assistance.
NOTES
*Corresponding author.