1. Introduction
E. coli is the most common pathogen of bacterial infections to man worldwide [1] . The presence of extended spectrum beta lactamase (ESBL) in clinical isolate has been documented as a very serious problem and a significant trait to: quick survival of patients in the hospital, high economic burden, lost of hours in life’s activities and high treatment failure. These ESBLs are enzymes that mediate resistance to extended-spectrum (third generation) cephalosporins (e.g., ceftazidime, cefotaxime, and ceftriaxone) and monobactams (e.g., aztreonam) but do not affect cephamycins (e.g., cefoxitin and cefotetan) or carbapenems (e.g., meropenem or imipenem) [2] . These bacterial enzymes have capacity to inactivate practically all cephalosporines [3] . Kenneth (2012) reported that ESBLs are plasmid mediated and the genes encoding these enzymes are easily transferable among different bacteria [4] . Most of these plasmids not only contain DNA encoding ESBLs but also carry genes conferring resistance to several non-β-lactam antibiotics. Other reports have also shown that plasmids in particular have been implicated in the spread of antibiotic resistance genes [5] . Enterobacteriaceae producing CTX-M β-lactamases have also shown typical resistant to quinolones, aminoglycosides and sulfonamides such as ciprofloxacine, gentamicin and trimethoprim/sulfamethoxazole, respectively [6] [7] . However, the detection of ESBLs genes can be difficult because they have different levels of activity against various antibiotics. Thus, the choice of antimicrobial agents to be used for clinical test is critical. For instance, an enzyme that would actively hydrolyze ceftazidime, to yield minimum inhibitory concentrations (MICs) of 256 µg/ml may produce MICs of 4 µg/ml on hydrolysis of cefotaxime. If an ESBL is detected, all penicillins, cephalosporins, and aztreonam should be reported as resistant, even if in vitro test results indicate susceptibility [2] [8] . However, multidrug resistance in Zaria metropolis can be attributed to multi-economic factors at the level of physician, patients, healthcare organizations and pharmaceutical companies, foster poor antibiotic usage i.e. behavioural and social environmental factors [9] . Based on these multi-economic factors that can trigger resistance to antibiotics in an environment, we proposed to revalidate the conventional method of ESBL analysis in clinical isolates using multiplex PCR technique.
2. Method
2.1. Isolation, Microscopy, Identification and Biochemical Test
Based on laboratory report, 10 multidrug resistant E. coli isolates were randomly collected from urine, stool, urogenital, wound and blood in A.B.U Teaching Hospital, Shika. Isolation, microscopy, identification and biochemical test of the E. coli isolates were carried out using standard microbiological methods [10] -[15] . Antibiotic susceptibility pattern of the isolates were carried out using disk diffusion method [16] . Interpretation of the susceptibility results were also carried out [17] . The antibiotics used were: Penicillin’s; ampicillin-sulbactam (SAM) (20 µg), amoxicillin (AML) (10 µg), amoxicillin-clavulanic acid (AMC) (30 µg). Cephalosporin’s: cefalexin (CL) (30 µg), ceftriazone (CRO) (30 µg), ceftazidine (CAZ) (20 µg), cefuroxime (CXM) (30). Fluoroquinolones: ciprofloxacin (CIP) (5 µg), nalidixic acid (NA) (30 µg), ofloxacin (OFX) (5 µg), Amino-glycosides: Gentamicin (CN) (10 µg). Tetracycline’s: tetracycline (30 µg). Miscellaneous agents: Chloramphenicol (C) (30 µg), Nitrofurantoin (F) (300 µg). The antibiotics were the commonly prescribed antibiotic at the Ahmadu Bello University, Teaching Hospital for the treatment of infections associated with E. coli.
2.2. Confirmatory Test for Extended Spectrum Beta Lactamases (ESBLS)
The double disc synergy test was adopted to detect the presence of ESBLs [18] . The identified and confirmed E. coli isolates with multidrug resistant characteristics were standardized in phosphate normal saline 5 ml using McFland 0.5 turbidity standard. The standardized organisms were streaked onto prepared Mueller Hinton agar and allowed to dry for 5mins at room temperature. Using a sterile pair of forceps, cefpodoxime (10 µg) and ceftriaxone (30 µg) disc were gently placed on the agar at a distance of 15 mm, center to center from a combination disc of amoxicillin-clavulanic acid (20:10 µg respectively). The plates were then incubated for 18 - 24 hrs and extended spectrum in the zone of inhibition was observed and interpreted. E. coli ATCC 25922 was used as a negative control. Positive result of ESBLs was interpreted as any isolate that has the zone around the test antibiotics disc increased towards the center disc of amoxicillin-clavulanic acid. The results were further interpreted using standard guidelines. A ≥5 mm increase in zone diameter for either antimicrobial agent compared to its zone when tested alone signifies positive result [17] .
2.3. Molecular Characterization of Resistant E. coli
2.3.1. Bacterial Culture Preparation
The bacteria cultures were prepared using a standard procedure [19] :
Luria and Bertani broth media were prepared (peptone, 10 g; NaCl, 5 g; 1N NaOH, 10 ml; yeast extract, 5 g; distill water 1 litre; pH 7.0 adjusted with NaOH solution) and sterilized at 121˚C for 15 mins. Single colonies were picked from isolates on MacConkey plate and inoculated into 5 ml Luria and Bertani (LB) broth medium and incubated overnight at 37˚C for 18 - 24 hrs. Bacteria culture was harvested by centrifugation at 4˚C, 8000 rpm (6800 × g) in a microcentrifuge for 30 seconds at room temperature in an Eppendorff’s tube. The supernatants were decanted and cells harvested.
2.3.2. Primers Used in This Study
The primers oligosequences used in the study of the selected resistance genes are shown in Table 1 below.
2.3.3. Genomic DNA Extraction
DNeasy extraction Kit (Qiagen, Germany) was used to isolate microbial genomic DNA from E. coli following manufacturer’s instruction [22] . The DNA was bound to silica gel membrane by passing the lysate through a column. Contaminants were washed away with the wash solutions (AW 1 and 2) and the DNA eluted with Manufacturer’s elution buffer. Purified DNA in the flow-through eluent was stored at −20˚C.
2.3.4. Agarose Gel Electrophoresis
Agarose gels were prepared and electrophoresis carried out [23] . 1% (w/v) agarose (Sigma) was dissolved in 1×
 (a)
(a)  (b)
(b)
Table 1 . Resistance genes and their primers oligosequences. Beta-lactams and cephalosporin primers used (a), Aminoglycoside Gentamicin primers used (b).
Accession no. (Reference or Source): AB083212 (46) [20] [21] .
TAE buffer by bringing to boil in a microwave oven. The gel was allowed to cool to about 40˚C before adding a drop (1 µg/ml) green nucleic acid gel stain which is used to replace the mutagenic ethidium bromide (EB). The gel was poured into a gel mold containing a well comb and allowed to polymerize at room temperature. Isolated DNA samples were mixed with 5 µl gel loading buffer and 20 µl of the sample was then loaded on to the wells of the gel. Electrophoresis was carried out at 100 mV for 45 minutes to allow easy separation of sample base on molecular weight. DNA bands were visualized and documented using an electrophoresis gel documentation system.
2.3.5. Amplification of Resistant Gene
Amplification of resistant gene; TEM, SHV and OXA was carried out in a 50 µl reaction mix. The master mix was prepared in a microtube comprising of; Dream Taq™ DNA polymerase (Fermenters) supplied with optimized 10× Dream Taq™ buffer, which includes 20 mM MgCl2, dNTP mix, template DNA (genomic DNA), and nucleasefree water. Enough master mix was prepared for the number of reactions plus one extra reaction to compensate for pipetting errors. The mix was then aliquot into thin-walled PCR tube earlier placed on an ice and the template DNA was added. The samples were vortexed gently and spun down before been transferred to the thermocycler. PCR was performed using the thermal cycling conditions below.
3. Results
Percentage Susceptibility of Multidrug Resistance Clinical Isolates of E. coli
The percentage antibiotic susceptibility profile of multidrug resistant clinical isolates of E. coli is as showed in the Figure 1 below.
The result showed that the isolates were 100% resistant to amoxicillin (AML), amoxicillin-clavulanic acid (AMC), cefuroxime (CXM) and ampicillin-sulbactan (SAM), 90% resistant to ceftazidine (CAZ) and tetracycline (TE), 70% resistant to ceftriazone (CRO), nalidixic acid (NA), cefalexin (CL) and ofloxacin (OFX), 60% to ciprofloxacin (CIP), 50% resistant to nitrofurantoin (F), 40 % resistant to chloramphenicol (C) and 20% resistant to gentamicin (CN).
Key: Ampicillin-sulbactam (SAM), Amoxicillin (AML), Amoxicillin-Clavulanic acid (AMC), Cefalexin (CL), Ceftriazone (CRO), Ceftazidine (CAZ), Cefuroxime (CXM), Ciprofloxacin (CIP), Nalidixic Acid (NA), Ofloxacin (OFX), Gentamicin (CN), Tetracycline (TE), Chloramphenicol (C), Nitrofurantoin (F), Multidrug resistance (MDR).
4. Production of Extended Spectrum β-Lactamase Evauation
The result of the test for the production of extended spectrum β-lactamase (ESBL) is as shown in Table 2.
This result showed that all the isolates were 100% resistant to amoxicillin-clavulanic acid, 70% to ceftazidime while 50% to ceftrazone and cefopodemin, with positive and negative phenotypic representation as shown in Figure 2. A ≥5 mm increase in zone diameter for either antibiotics compared to its zone when tested alone signified a positive ESBL.
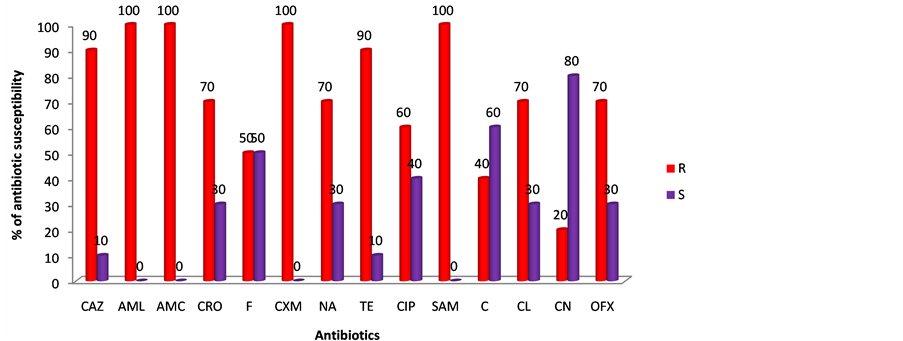
Figure 1. Percentage susceptibility of multidrug resistance clinical isolates of E. coli.
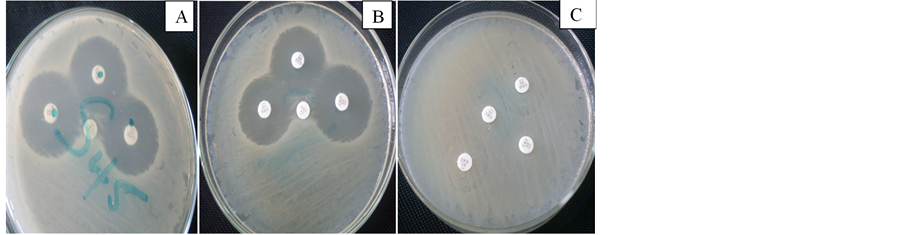
Figure 2. Pictorial representation of expressed extended spectrum β-lactamase producing isolates of E. coli using Double Disc Diffusion Method.

Table 2. Determination of extended spectrum β-lactamase (ESBL) using double disc diffusion method.
Production of Extended Spectrum β-Lactamase Using Double Disc Diffusion Method
The evaluation of microbial production of extended spectrum β-lactamase using double disc diffusion method is as shown below.
Picture A and B (positive result) showed the expressed extended spectrum β-lactamase enzyme through diffusion while C showed no diffusion (negative result).
5. Molecular Characterization of Resistant Gene of E. coli Isolates
The result of the genomic DNA extraction on agarose gel electrophoresis is shown in Figure 3.
The above result in Figure 3 showed a DNA ladder base pair in ng/0.5 µg, 8 cm length gel and 1.0% agarose gel electrophoresis (X1 TAE, 7 v, 30 min). Genomic DNA isolated from 9 multiple drug resistant E. coli isolates from different clinical samples in A.B.U Teaching Hospital Shika, Zaria. Isolate U60 was not extracted because of shortage in reagent.
Lane 1: Genomic DNA from E. coli ATCC 25922 as control.
Lane 2: Genomic DNA multidrug resistant clinical isolate W15.
Lane 3: Genomic DNA from multidrug resistant clinical isolate B2.
Lane 4: Genomic DNA from multidrug resistant clinical isolate B3.
Lane 5: Genomic DNA from multidrug resistant clinical isolate U64.
Lane 6: Genomic DNA from multidrug resistant clinical isolate S45.
Lane 7: Genomic DNA from multidrug resistant clinical isolate U58.
Lane 8: Genomic DNA from multidrug resistant clinical isolate S57.
Lane 9: Genomic DNA from multidrug resistant clinical isolate B7.
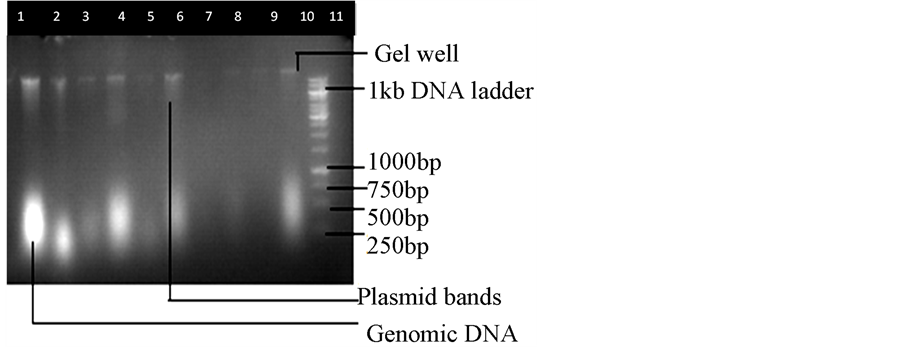
Figure 3. Genomic DNA extraction on agarose gel electrophoresis.
Lane 10: Genomic DNA from multidrug resistant clinical isolate URO2.
Lane 11: 10,000 base pair of the genomic DNA ladder.
Multiplex PCR Genomic DNA with Primer Annealing on Gel Electrophoresis
The result of the multiplex PCR analysis is as shown in Figure 4.
6. From the Left to the Right Bands
Lane 1: 10 kbp plus genomic DNA ladder composing of DNA fragments in base pairs: 10,000, 8000, 6000, 5000, 4000, 3500, 3000, 2500, 2000, 1500, 1000, 750, 500 and 250.
Lane 3: isolate W15 showed three (3) bands of resistant gene, which is assumed to be TEM with base pair 931, SHV with base pair 868 and OXA-2 (478). This result was interpreted using the manufacturer DNA ladder base pair bands.
Lane 5: isolate B2 showed only a band of resistant gene, which is interpreted to be OXA-2 with base pair 478.
Lane 7: isolate URO2 showed 2 bands of SHV base pair 868 and OXA-2 with band of 478.
Lane 8: isolate U64 showed 2 bands of TEM and OXA-2 with base pair 478.
Lane 9: isolate S45 showed 2 bands TEM and OXA with base pair 931 and 478 respectively.
Lane 2, 4 and 6: isolates S57, U58, B7 showed no bands respectively but they show various degree of resistant in MIC and antibiotic susceptibility profile test.
7. Discussion and Conclusion
Extended spectrum β-lactamase indeed is a superbug of trouble to clinicians, creating environmental stress to pharmaceutical pipeline in the development of new antibiotics. The antibiotic susceptibility profile of the multidrug resistance isolates with ESBLs as showed in Figure 1, showed that 100% of the isolates were resistant to amoxicillin-clavulanic acid, amoxicillin, cefuroxime and ampicillin-sulbactam, 90% to ceftazidine and tetracycline, 70% to ceftriazon, ofloxacin, nalidixic acid and cefalexin, 60% to ciprofloxacin, 50% to nitrofurantoin, 40% to chloramphenicol and 20% to gentamicine. Suggesting that bacteria with ESBL enzymes have capacity to inactivate practically all cephalosporines [3] as CTX-M β-lactamases producing Enterobacteriaceae are typically resistant to quinolones, aminoglycosides and sulfonamides such as ciprofloxacine, gentamicin and trimethoprim/ sulfamethoxazole [6] [7] . A report in Lagos has suggested that E. coli could show maximum sensitivity to ceftriazon and ciprofloxacin (at mean score of 85) while gentamycin might have broad spectrum activity due to limited use [24] [25] . Also findings in Enugu using E. coli from livestock suggested that E. coli isolates can express resistance rates of 85% to ampicillin; 22.5% to ceftriaxone; 18.8% to cefoxitin; 16.3% to nalidixic acid and 12.5% to gentamycin [26] , but the variation in rate of resistant of microorganism to antibiotic might be a function of misuse of antibiotics in such location as shown in the high multidrug resistance index of 0.6 and above in Table 3, indicating that these isolates which are from antibiotic sources have been widely used [27] .

Table 3. Multiple antibiotic resistant Index of the multidrug resistant isolates.
Key: Ampicillin-sulbactam (SAM), Amoxicillin (AML), Amoxicillin-Clavulanic acid (AMC), Cefalexin (CL), Ceftriazone (CRO), Ceftazidine (CAZ), Cefuroxime (CXM), Ciprofloxacin (CIP), Nalidixic Acid (NA), Ofloxacin (OFX), Gentamicin (CN), Tetracycline (TE), Chloramphenicol (C), Nitrofurantoin (F), Multidrug resistance (MDR).
Other reports suggest that ESBLs in gram-negative bacteria have been implicated as the enzymes responsible for resistance to β-lactam antibiotics such as cefotaxime, ceftazidime and aztreonam [28] -[30] . Since these enzymes were first identified in Western Europe, more than 70 ESBLs have been found worldwide including Nigeria [30] . Plasmid-encoded class A TEM and SHV type enzymes ESBLs evolution are attributed to successive mutations in their structural genes, resulting in either single or multiple amino acid changes in the encoded enzymes [31] . In this work, determination of extended spectrum β-lactamase resistance using double disc diffusion method as shown in Table 2, showed that out of the 10 isolates observed, all the isolates showed 100% resistance to amoxicillin-clavulanic acid, 70% to ceftazidime, 50% to ceftrazone and cefopodemin. The positive and negative phenotypic representations of the ESBL isolates in E. coli are as shown in Figure 2. Seventy percent (70% (7/10)) (Table 2) of the selected multidrug resistant clinical isolates showed ESBLs positive; a ≥5mm increase in zone diameter for either antibiotics compared to its zone when tested alone. The isolates producing ESBL were further evaluated for ESBLs gene (OXA, SHV and TEM) using multiplex PCR technique as shown in Figure 4 above. The genomic DNA extraction (Figure 3) result showed that 8 of the multidrug resistant isolates genomic DNA were extracted, which showed pronounced glowing at base pair of 1000 below on gel electrophoresis, one of the isolates did not showed glowing trace of DNA, while well 1 (B7) showed faint glowing. These defects could be attributed to over homogenizing of the cells with gel beads and/or cell lyser in vortexing machine. The multiplex PCR ran at the thermocycling conditions as shown in Table 4, with primers TEM (931 bp), SHV (868), OXA-2 (478), aac(3)-IIa (900) and rmtA (634) (Table 1), which are gene responsible for ex

Table 4. Thermal cycling conditions for resistant gene application. Primers conditions: TEM (931), SHV (868), OXA-2 (478), aac(3)-IIa (900), rmtA(634).
Total time is 2 hrs 19 mins.
tended spectrum β-lactamase and aminoglycoside resistance in E. coli showed that isolate W15 comprises of three (3) resistant gene, which corresponded with TEM resolving at 931 base pair, SHV 868 base pair, and a 478 bp indicating OXA-2 that is faint probably indicating a low concentration of the gene, isolate B2 comprises single resistant gene, which is interpreted as OXA-2 with 478 base pair, isolate URO2, U64 and S45 comprises of two resistance genes which resolve as 868 and 478 base pair indicating SHV and OXA-2 respectively. However, isolates S57, U58 and B7 showed no gene amplification despite the various degree of resistance in MIC and antibiotic susceptibility profile test obtained with conventional detection analysis (Table 3). We assume that their resistant genes are not coded for by the primers used in this study as these isolates are likely to contain other resistant genes, which are also expressed at a molecular level. However, reports have justified that isolates resistant to gentamicin, kanamycin, or streptomycin can show negative bands for all genes tested [ant(6)-Ia, ant(9)-Ia, ant(49)-Ia, aph(39)-IIIa, aph(20)-Ib, aph(20)-Ic, aph(20)-Id, aac(69)-Ie-aph(20)-Ia, and aac(69)-Ii], indicating that additional resistance genes might exist but not expressed in molecular level [32] while Nadine et al.’s (2012) findings correlate with our report that more than two ESBLs genes can be isolated in a single isolate using multiplex PCR technique [33] [34] . In validating this work, the multiplex PCR result showed a confirmation of the double disc diffusion test and this justifies the claim that the resistant genes are present. The detections of TEM, SHV, OXA encoding genes for resistance to ESBLs were reported to be plasmid encoded implying that these resistance determinants are found in our environment and can be transferred from one organism to another. These resistant traits call for gross surveillance in clinical settings because ESBLs gene is a world growing trait for available antibiotics, but its epidemiological effect and evaluation are still underestimated with low awareness.
Abbreviations
ESBLs = Extended Spectrum Beta-Lactamases TEM = Temoneira SHV = Sulfhydryl Variable OXA = Oxacillin PCR = Polymerase Chain Reaction MIC = Minimum Inhibition Concentration CDC = Centre for Disease Control and Prevention DNA = Deoxyribonucleic Acid CTX-M = Cefotaxime ABUTH = Ahmadu Bello University Teaching Hospital ATCC = American Type Culture Collection CLSI = Clinical and Laboratory Standards Institute LB = Luria and Bertani F = Forward R= Reverse Bp = Base Pair U = Isolate from Urine URO = Isolate from Urogenital S = Isolate from Stool B = isolate from Blood
NOTES
*Corresponding author.