Automated Classification of Lung Diseases in Computed Tomography Images Using a Wavelet Based Convolutional Neural Network ()
1. Introduction
Lung cancer is the deadliest cancer in the world with 1.69 million deaths in 2015 alone, according to the World Health Organization. Early detection by CT screening can decrease lung cancer mortality. However, lung cancer screening is time consuming and costly. To deal with this issue, development of methods for automatic detection of pulmonary lesions and automatic classification of pulmonary diseases in digital chest radiographs and computed tomography (CT) scans has been an active area of research for the last two decades [ 1 - 4 ]. In recent years, the number of lung cancer screening using CT images has increased, and computer-aided diagnosis (CAD) systems play a vital role in the interpretation. So far, numerous studies on the development of CAD systems for lung cancer in CT have been reported [ 5 - 8 ]. Many CAD systems have been developed based on extracting features from images. The features are then discriminated using various classifiers, such as linear discriminant analysis and support vector machine (SVM). Most traditional CAD systems mainly relay on manually designed features. However, the features extracted from images may not be optimal or adequate. Thus, it is still a significant challenge to further improve the performance of CAD systems [ 9 ].
Recently, deep learning has attracted much attention in many fields, such as image recognition and biomedical image analysis. Convolutional neural network (CNN) is one of the most popular algorithms for deep learning. CNN has been successfully applied to various fields and has achieved state-of-the-art performance in image recognition and classification [ 10 , 11 ]. After its success, CNN is also exploited in medical fields, such as image processing and CAD [ 12 - 15 ]. Of these studies, some researches on pulmonary diseases have been conducted [ 16 - 19 ]. In these studies, interstitial lung disease pattern detection [ 16 , 17 ], automatic detection of pulmonary nodules [ 18 ], and detection of ground glass opacity regions [ 19 ] using CNN are discussed. But there is still room for improvement on performance. We believe that enhancing invariance of image features is a way to improve the performance.
In this study, we aim to construct an automatic system using a wavelet transform based CNN for lung cancer classification. Normally, CNN performs image classification directly on raw image pixels and it is implemented in the spatial domain. However, in this case, the spectral information content of the image is not utilized in the classification. We consider that further improve image classification performance can be achieved by incorporating spectral feature information for enhancing invariance of image features.
In this paper, we propose a method for automatic classification of pulmonary diseases by using wavelet coefficients as inputs to CNN. We classify four types of lung cancer in CT images including normal case, into four categories, i.e., lung adenocarcinoma, lung squamous cell carcinoma, metastatic lung cancer, and normal. In order to demonstrate the effectiveness of the proposed method, we compare and evaluate a conventional transfer learning-based method and a SVM classification method.
2. METHODS
The neocognitron proposed by Fukushima in 1980 [ 20 ] could be regarded as the predecessor of CNN. It is a multi-layered convolutional network that can be trained to recognize visual patterns. A basic CNN consists of three components: convolutional layer, pooling layer, and output layer. With the multilayer structure by cascading these layers, the dimension of the pattern data of the input image gradually decreases. Finally, the features of input image are mapped to the output layer (fully connected layer) for classification. In this study, we utilize AlexNet [ 21 ] which is a well-known CNN-based model for classification. AlexNet has showed excellent performance in general image recognition and won the 2012 ImageNet [ 22 ] LSVRC-2012 competition. For constructing our proposed network, we utilized the first half part the pre-trained AlexNet (Hereinafter referred to as the pre-trained network). And then we newly designed the second half part (un-trained fully connected layers) for our proposed network. With this network, transfer learning and fine tuning were performed. At the re-training phase, the wavelet coefficients obtained from the images were used as the input to the network. Region of interest (ROI) for suspected lesions in images was not provided. That is, the inputs for the proposed network are the wavelet coefficients of the entire image. After re-training, we classify the lung CT images into four categories using the proposed network.
2.1. Image Data Set
The data set used in this study is DICOM images of lung CT scans obtained from a database published on the Internet (The Cancer Imaging Archive) [ 23 ]. Therefore, approval from an ethics committee and informed consent are not required. Our dataset contains 448 cases including 100 cases of lung adenocarcinoma (LUAD), 125 cases of lung squamous cell carcinoma (LUSC), 82 cases of metastatic lung cancer (meta), and 141 cases of normal (normal). All of the confirmed cases were diagnosed by experienced radiologists. Each of the 4 categories was obtained from 25 patients. From each case, we selected 3 to 4 slices containing suspected lesions. As an example, Figure 1 shows 4 images representing the 4 respective categories. The basic information about 448 lung cases is summarized in Table 1.
2.2. Wavelet Transforms
Multiresolution analysis using the wavelet transform (WT) has received considerable attention in the past two decades in various fields. In the medical field, two-dimensional WT has been applied to data compression, image enhancement, noise removal, etc. [ 24 ]. In the multiresolution analysis, an image is initialized at level 0 (original image without decomposition). The image is decomposed into four components of level 1 (one low frequency component and three high frequency components). A smoothed image can be obtained from the low frequency component, and detailed images, such as directional edges can be obtained from the three high frequency components. Therefore, low frequency component is also referred to as smoothed component, and high frequency components referred to as detailed components. Decomposition is further performed on the low frequency component. When the decomposition is repeated in such a way, the resolution level becomes higher, and the resolution of the image decreases as the level increases. For the details of the WT, refer to the literature by Daubechies [ 25 ].
When WT is implemented, down sampling is usually performed. Thus, resolution level increases by 1, and the sample sizes of the four decomposition components become 1/4, respectively. The decimated WT affects shift invariance and causes problems, such as disappearance of the outline of the decomposed images.
In order to overcome this issue, we used the redundant discrete wavelet transform (RDWT). Unlike the WT, the RDWT does not incorporate the down sampling operations. Thus, the low-frequency coefficient and the high-frequency coefficients at each level are the same length as the original signal. The basic
![]()
Figure 1. Lung CT dataset sample. (a) adenocarcinoma (LUAD). (b) squamous cell carcinoma (LUSC), (c) metastatic lung cancer (meta), and (d) normal.
![]()
Table 1. The basic information about our data set.
algorithm of the conventional RDWT is that it applies the transform at each point of the image and saves the detailed coefficients and uses the approximation coefficients for the next level. The size of the coefficients array does not diminish from level to level [ 24 ].
The wavelet basis function used in the present study was Daubechies order 2 (db2). Figure 2 shows an original image and the visualized images of 4 decomposed components of the original image at level 1.
2.3. Architecture of the Proposed CNN
CNN training is implemented with the Caffe deep learning framework [ 26 ], using MacOS 10.13.4, Processor; 1.8 GHz, Intel Corei 5, Graphics; Intel HD Graphics 4000, Interface; MATLAB. Input data to the network is the wavelet coefficients obtained from original lung CT images. The original image (pixel value) of the lung CT is also used as inputs for comparison and verification. Wavelet coefficients used as input data are one low-frequency smoothed component (low-low component: LL) and three high-frequency detailed components (low-high component: LH, high-low component: HL, high-high: HH component). We combined three of the four components and used them as inputs to the network. Five combinations which have shown better performance in our pre-test were selected. Figure 3 shows the outline of the combinations used for the three channel input.
At the first step, we extract out the first-half-part layer of AlexNet’s pre-trained network, which is the
![]()
Figure 2. Original image and four decomposed images at level 1, obtained by redundant discrete wavelet transform using Daubechies db2 basis function. (a) Original image. (b) LL component. (c) LH component. (d) HL component. (e) HH component.
![]()
Figure 3. Level 1 wavelet decomposition of an original image. (a) Combination of three identical original images. (b) Combination of LL, LH and HL components. (c) Combination of LL and two HH components. (d) Combination of three LL components. (e) Combination of two LL and one HH components. (f) Combination of LH, HL and HH components.
feature expression part. In the second step, the acquired feature expression part is then connected to a newly designed second half part (un-trained fully connected layers). It is the basic architecture of network that we proposed. We applied 50% dropout among the fully connected layers to randomly deactivate the units for learning. By doing so, the weights are changed at the end of each training iteration. It results in enhancing generalization performance. Additionally, L2-norm regularization is adopted for preventing overfitting and for enhancing generalization performance. In the third step, the wavelet coefficients of the 3 component images are inputted to the three-input channels of the newly constructed network, followed by performing back propagation for re-training. Figure 4 shows the flow chart of the proposed method. Figure 4(a) is the pre-trained AlexNet, and Figure 4(b) is the basic architecture of the proposed method.
We used the stochastic gradient descent method with momentum of 0.9 to train the network. Approximately 70% (314 cases) of the data set (448 cases) were used for training and the remaining 30% (134 cases) for verification. To determine the number of epochs, accuracy validation was performed at the end of each iteration. The learning process stops, if the value of accuracy does not reach to the maximum after 5 iterations. The cross entropy was used as a loss function.
The traces of the training loss and accuracy of the proposed network are shown in Figure 5. There are three graphs in the figure. Learn represents the loss of each mini batch, Learn (smoothed) stands for the loss been smoothed (moving average of each mini batch), and Validation is a curve representing the loss per iterative learning. In Figure 5(a), training loss significantly decreases with the increase of the number of iterations. Figure 5(b) shows the relationship between mini batch classification accuracy and the number of iterations. It is obvious that accuracy increases with the increase of the number of iterations. From the results of Figure 5, it is reasonable to assume that learning has been appropriately implemented.
We used holdout method for cross verification. Out of the 448 images, approximately 70% (314 cases) were used for training and the remaining 30% (134 cases) for verification. In order to demonstrate the superiority of the proposed method, a method of re-training the pre-trained network without the replacement of the fully-connected layers (hereinafter referred to as transfer learning), and a method of using pre-trained network with SVM classifier instead of the fully-connected layers (hereinafter referred to as CNN + SVM) were also used for comparison.
![]()
Figure 4. Flowchart of the proposed method for classification of lung diseases in CT images. (a) AlexNet. (b) Proposed network.
3. RESULTS
In our proposed method we replaced the 2 fully-connected layers of AlexNet with new layers to construct a new network for classification task. The input data used were the redundant wavelet coefficient of the target images. For comparison, two methods, namely CNN + SVM method and existing transfer learning method were also used to classify the lung diseases in CT scans. The classification accuracy rates for lung disease detection using the three methods are shown in Table 2. The values in the leftmost column
![]() (a)
(a)![]() (b)
(b)
Figure 5. Traces of loss and accuracy during training. (a) Relationship between loss and iteration. Loss refers to the difference from correct answer. (b) Relationship between accuracy and iteration.
![]()
Table 2. Classification results obtained using the three methods: CNN+SVM method, CNN transfer learning method, and the proposed method.
of the table are overall accuracy. The overall accuracy for the five combinations of three wavelet coefficients used as 3-channel inputs are listed. Other than the overall accuracy, the values in the table are average values obtained with 10 measurements. Student’s t-test was used for statistical analysis. Significance levels used were 0.05 and 0.01.
A confusion matrix is usually used as the quantitative method of characterizing image classification accuracy. The confusion matrix indicates the correspondence between the predicted result and a ground truth. The three confusion matrices shown in Table 3 were used for comparison of classification accuracy for CNN+SVM method, conventional transfer learning method and the proposed method using LL, LH and HL wavelet coefficients as inputs. Precision and recall shown in the figure are used as evaluation indices obtained from the confusion matrix.
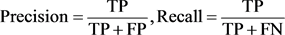
TP: true positive, FP: false positive, FN: false negative.
Figure 6 shows the receiver operating characteristic (ROC) curves and the area under the curve (AUC) of the four categories obtained from the proposed method and the other two methods.
4. DISCUSSION
It is obvious from Table 2 that our proposed method shows the highest overall accuracy as compared to the CNN+SVM method and the transfer learning method. For example, when using the combination of one low-frequency component (LL) and two high-frequency components (LH and HL) as inputs to the proposed network, an overall accuracy of 0.919 is achieved. While the overall accuracy of the conventional transfer learning method (without the replacement of the full-connected layers) by inputting the pixel
Table 3. Confusion matrices obtained from three various classifiers. (a) CNN + SVM (original * 3), (b) CNN with transfer learning, and (c) the proposed method using LL, LH, HL wavelet coefficients.
values of the original images to the input channels is 0.874, and that of CNN+SVM is 0.844. A statistically significant difference between the proposed method and the CNN + SVM method (p < 0.01) exists. There is also a statistically significant difference (p < 0.05) between the proposed method using LL, LH, HL wavelet coefficients and the conventional transfer learning method. As for the other combinations of wavelet coefficients, the proposed method shows higher classification accuracy than the transfer learning method, although the statistically significant difference is not found. However, the proposed method has the tendency to complete the learning course with fewer training epochs. In other words, compared to the transfer learning method, the proposed method provides fast convergence, suggesting that the proposed method has equal or higher accuracy with fewer number of learning. From these results, we believe that the proposed method is superior to the transfer learning method in terms of computing efficiency. As shown in Table 2, in the case that wavelet coefficients are used as inputs, the classification accuracy rates achieve 0.9 or higher in any of the five combinations. In particular, when the input includes the low-frequency component LL, higher classification accuracy can be obtained. This may be because the low frequency component contains a relatively large amount of original image information.
As shown in Table 3, recall of the proposed method is significantly higher than the other two methods for LUAD classification. Namely, the proposed method can correctly classify LUAD more precisely than the other two methods. However, the proposed method only achieves an 85% precision and is comparable to the transfer learning method. Generally speaking, this classification performance is not high enough, although it is higher as compared to the CNN + SVM method (p < 0.05). As for LUSC, precision of the proposed method is significantly higher than the other two methods. Regarding metastatic lung cancer and normal cases, the proposed method also shows the highest accuracy. These results demonstrate the superiority of the proposed method.
From the shape of the ROC curves and the values of AUC shown in Figure 6, the proposed method has higher performance than any of the other two methods for all of classifications. These results suggest that the proposed method outperforms the other two methods. Another advantage of the proposed method is that it is not necessary to extract the suspected lesion areas in advance. Instead of extracting ROI, the proposed method uses the wavelet coefficients of the whole image. In order to further demonstrate our proposed method, the precision, recall and F1-measure of the three methods are summarized in Table 4. It is obvious from the table that our proposed method outperforms the other two methods.
The proposed method could not achieve 100% accuracy due to misrecognition, thus there is ample room for further improvement. In this study, the wavelet basis function used is Daubechies order 2 (db2). It is undeniable that the use of other basis functions may lead to better results. We only used AlexNet for the present study. We plan to utilize other CNN architectures, such as ResNet and VGG for comparison in our future work.
5. CONCLUSIONS
In this paper we proposed a novel transfer learning method that utilized the pre-trained network based on AlexNet and connected it to the newly designed fully-connected layers. We used wavelet coefficients as inputs for classifying four types of lung cancer including normal cases from lung CT images. To validate the usefulness and effectiveness of the proposed method, we also evaluated a conventional transfer learning method and a SVM-based method. Our proposed method achieves overall accuracy of 91.9% when LL, HL, and LH components were used. The overall accuracy is higher than that obtained by the other two methods. This demonstrates the superiority of the proposed method. Another advantage of the
![]()
Table 4. Comparison of the performance of the three methods in terms of precision, recall and F1-measure.
proposed method is that it is unnecessary to extract the suspected lesion areas in advance. Instead of the ROI, the wavelet coefficients of the whole image were used in the proposed method.
In future studies, in order to further improve classification accuracy we plan to investigate the effect on classification performance by selecting wavelet basis functions other than db 2 and pursue to design a new architecture. We believe that the proposed method can be used in other disease classification and different imaging modalities.
Acknowledgements
This work was supported in part by JSPS KAKENHI Grant Number 18K15641, and in part by a grant from the International University of Health and Welfare.