A Comparison of Two Linear Discriminant Analysis Methods That Use Block Monotone Missing Training Data ()
Received 7 January 2016; accepted 23 February 2016; published 26 February 2016

1. Introduction
We consider the problem of classifying an unlabeled observation vector 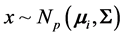 into one of two distinct multivariate normally distributed populations
into one of two distinct multivariate normally distributed populations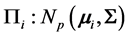 ,
, 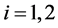 , when monotone missing training data are present, where
, when monotone missing training data are present, where 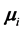 and
and 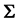 are the
are the 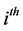 population mean vector and common covariance matrix, respec- tively. Here, we re-compare two linear classification procedures for block monotone missing (BMM) training data: one classifier is from [1] , and the other classifier employs the maximum likelihood estimator (MLE).
population mean vector and common covariance matrix, respec- tively. Here, we re-compare two linear classification procedures for block monotone missing (BMM) training data: one classifier is from [1] , and the other classifier employs the maximum likelihood estimator (MLE).
Monotone missing data occur for an observation vector 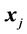 when, if
when, if 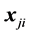 is missing, then
is missing, then 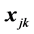 is missing for all
is missing for all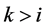 . The authors [1] claim that their “linear combination classification procedure is better than the substitution methods (MLE) as the proportion of missing observations gets larger” when block monotone missing data are present in the training data. Specifically, [1] has performed a Monte Carlo simulation and has concluded that their classifier performs better in terms of the expected error rate (EER) than the MLE sub- stitution (MLES) classifier formulated by [2] as the proportion of missing observations increases. However, we demonstrate that for intra-class covariance training data with at least small correlations among the variables, the MLES classifier can significantly outperform the classifier from [1] , which we refer to as the C-H classifier, in terms of their respective EERs. This phenomenon occurs regardless of the proportion of the variables missing in each observation with missing data (POMD) in the training data set.
. The authors [1] claim that their “linear combination classification procedure is better than the substitution methods (MLE) as the proportion of missing observations gets larger” when block monotone missing data are present in the training data. Specifically, [1] has performed a Monte Carlo simulation and has concluded that their classifier performs better in terms of the expected error rate (EER) than the MLE sub- stitution (MLES) classifier formulated by [2] as the proportion of missing observations increases. However, we demonstrate that for intra-class covariance training data with at least small correlations among the variables, the MLES classifier can significantly outperform the classifier from [1] , which we refer to as the C-H classifier, in terms of their respective EERs. This phenomenon occurs regardless of the proportion of the variables missing in each observation with missing data (POMD) in the training data set.
Throughout the remainder of the paper, we use the notation  to represent the matrix space of all
to represent the matrix space of all 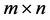 matrices over the real field
matrices over the real field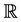 . Also, we let the symbol
. Also, we let the symbol 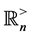 represent the cone of all
represent the cone of all 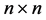 positive definite matrices in
positive definite matrices in![]() . Moreover,
. Moreover, ![]() represents the transpose of
represents the transpose of![]() .
.
The author [3] has considered the problem of missing values in discriminant analysis where the dimension and the training-sample sizes are very large. Additionally, [4] has examined the probability of correct classi- fication for several methods of handling data values that are missing at random and use the EER as the criterion to weigh the relative quality of supervised classification methods. Moreover, [5] has examined missing obser- vations in statistical discrimination for a variety of population covariance matrices. Also, [6] has applied re- cursive methods for handling incomplete data and has verified asymptotic properties for the recursive methods.
We have organized the remainder of the paper as follows. In Section 2, we describe the C-H classifier, and we describe the MLES linear discriminant procedure when the training data from both classes contain identical BMM data patterns. In Section 3, we describe and report the result of Monte Carlo simulations that examine the differences in the estimated EERs of the C-H and MLES classifiers for various parameter configurations, training-sample sizes, and missing data sizes and summarize our simulation results graphically. In Section 4, we compare the C-H and MLES linear classifiers using a parametric bootstrap estimator of the EER difference (EERD) on two actual data sets. We summarize our results and conclude with some brief comments in Section 5.
2. Two Competing Classifiers for BMM Training Data
2.1. The C-H Classifier for Monotone Missing Data
Suppose we have two ![]() training observation matrices in the form
training observation matrices in the form
![]() (1)
(1)
where
![]() (2)
(2)
denotes the ![]() complete-observation submatrix, and
complete-observation submatrix, and ![]() is the partial observation submatrix whose first k measurements are non-missing, where
is the partial observation submatrix whose first k measurements are non-missing, where![]() , for
, for![]() . We denote a complete data ob- servation vector by
. We denote a complete data ob- servation vector by![]() , where
, where ![]() and
and ![]() such that
such that
![]() (3)
(3)
where![]() ,
, ![]() ,
, ![]() ,
, ![]() , and
, and ![]() with
with ![]() . Also, random samples of sizes
. Also, random samples of sizes ![]() are taken from distributions of the form
are taken from distributions of the form![]() , where
, where ![]() and
and![]() .
.
The authors [1] have derived a linear combination of a discriminant function composed from complete data and a second discriminant function determined from BMM data. The C-H classifier uses Anderson’s linear dis- criminant function (LDF) for the subset of complete data![]() ,
, ![]() , given in (2), which is
, given in (2), which is
![]()
where ![]() and
and
![]()
are the complete-data sample mean and complete-data sample covariance matrix, respectively. They also use Anderson’s LDF for the data
![]() (4)
(4)
![]() , with k features and
, with k features and ![]() training observations, which is
training observations, which is
![]()
with
![]() (5)
(5)
where
![]() (6)
(6)
denotes the sample mean for the first ![]() observations and the first k features from
observations and the first k features from ![]() in (1),
in (1),
![]()
denotes the sample mean for the first k features of the latter ![]() observations from
observations from ![]() in (1), and
in (1), and
![]()
is the pooled sample covariance matrix for the incomplete training data (4), where![]() , represent the subsets of (1) with non-missing data and BMM data, respectively, for
, represent the subsets of (1) with non-missing data and BMM data, respectively, for![]() .
.
The authors [1] have proposed the linear combination statistic
![]() (7)
(7)
where![]() . One classifies an unlabeled observation vector
. One classifies an unlabeled observation vector ![]() into
into ![]() if
if
![]() (8)
(8)
and into![]() , otherwise. The conditional error rate (CER) for classifying an unlabeled vector x from
, otherwise. The conditional error rate (CER) for classifying an unlabeled vector x from ![]() into
into ![]() using (8) is
using (8) is
![]() (9)
(9)
![]() ,
, ![]() , where
, where
![]() (10)
(10)
with
![]()
Also,
![]() (11)
(11)
where ![]() and
and![]() ,
, ![]() ,
, ![]() ,
, ![]() ,
, ![]()
![]() , with
, with![]() ,
, ![]() , and
, and ![]() defined in (5). Thus, using (9) and assuming equal a priori probabilities, the CER for (8) is
defined in (5). Thus, using (9) and assuming equal a priori probabilities, the CER for (8) is
![]() (12)
(12)
If
![]()
then, for (8), the EER of misclassifying an unlabeled observation vector ![]() from
from ![]() into
into ![]() is
is
![]()
![]() ,
,![]() . Thus, once again assuming equal a priori probabilities, the EER for (8) is
. Thus, once again assuming equal a priori probabilities, the EER for (8) is
![]()
In choosing c in (7), [1] have utilized the fact that the CER and EER will depend on the Mahalanobis distance for the complete and partial training observations and the corresponding training-sample sizes, ![]() and
and![]() ,
,![]() . Usually, when one has small CERs, at least one of the sample Mahalanobis distances
. Usually, when one has small CERs, at least one of the sample Mahalanobis distances
![]()
will be large. While ![]() and
and ![]() determine the performance of
determine the performance of![]() , the quantities
, the quantities ![]() and
and ![]() dictate the performance of
dictate the performance of![]() . Hence, [1] have chosen c in relation to the training-sample sizes and the Mahalanobis dis- tances for the complete and incomplete training-data sets. Note that the implication for circumstances where
. Hence, [1] have chosen c in relation to the training-sample sizes and the Mahalanobis dis- tances for the complete and incomplete training-data sets. Note that the implication for circumstances where ![]() is that the information in the data-matrix component
is that the information in the data-matrix component![]() ,
, ![]() , in (1) contributes largely to the discriminatory information. Hence, [1] uses
, in (1) contributes largely to the discriminatory information. Hence, [1] uses
![]()
to determine the linear combination classification statistic (7).
2.2. A Maximum Likelihood Substitution Classifier for Monotone Missing Training Data
The authors [7] have derived an MLE method for estimating parameters in a multivariate normal distribution with BMM data. The estimator of ![]() in the [7] MLES classifier is a pooled estimator of the two individual MLEs of
in the [7] MLES classifier is a pooled estimator of the two individual MLEs of![]() .
.
Below, we state the MLEs for two multivariate normal distributions having unequal means and a common covariance matrix with identical BMM-data patterns in both training samples.
Theorem. Let ![]() be modeled by the multivariate normal densities
be modeled by the multivariate normal densities ![]() for
for![]() , with
, with
![]() (13)
(13)
and
![]() (14)
(14)
Also, let
![]()
![]()
![]()
and
![]()
where![]() , and
, and ![]() with
with![]() ,
, ![]() , and
, and ![]() given in (1). Then, on the basis of two-step mono- tone training samples from populations
given in (1). Then, on the basis of two-step mono- tone training samples from populations![]() , the MLEs of (13) and (14) are
, the MLEs of (13) and (14) are
![]() (15)
(15)
respectively, where
![]()
![]() (16)
(16)
and
![]() (17)
(17)
with![]() , where
, where ![]() is defined in (5),
is defined in (5),
![]()
![]()
and
![]()
where![]() ,
, ![]() ,
, ![]() , and
, and![]() , are defined in (5), (6), (16), and (17), respectively, for
, are defined in (5), (6), (16), and (17), respectively, for![]() .
.
Proof: A proof is alluded to in [8] .
The MLES classification statistic is
![]() (18)
(18)
where![]() ,
, ![]() , and
, and ![]() are the MLEs defined in (15), and
are the MLEs defined in (15), and ![]() is an unlabeled observation vector belonging to either
is an unlabeled observation vector belonging to either ![]() or
or![]() . We classify the unlabeled observation vector
. We classify the unlabeled observation vector ![]() into
into ![]() if
if
![]() (19)
(19)
and into![]() , otherwise. Given that
, otherwise. Given that![]() , conditioning on
, conditioning on![]() ,
, ![]() , and
, and![]() , and using the fact that
, and using the fact that
![]()
where![]() , along with (15), (18), and (19), we have that
, along with (15), (18), and (19), we have that
![]()
where
![]() (20)
(20)
![]() . Similarly, given
. Similarly, given![]() ,
,
![]()
where ![]() is given in (20). Thus, assuming equal a priori probabilities of belonging to
is given in (20). Thus, assuming equal a priori probabilities of belonging to![]() ,
, ![]() , for an unlabeled observation, we have
, for an unlabeled observation, we have
![]() (21)
(21)
Hence, the overall expected error rate is
![]()
3. Monte Carlo Simulations
The authors [1] claim that “it can be shown that the linear combination classification statistic is invariant under nonsingular linear transformations when the data contain missing observations” and assume this invariance is also true for the MLES classifier. While their assertion might be true for the C-H classifier, it is not necessarily true for the MLE classifier. Because [1] do not consider covariance structures with moderate to high correlation, their results are biased toward the C-H classifier. Here, we show that the MLES classifier can considerably outperform the C-H classifier, depending on the degree of correlation among the variables with missing data and the variables without missing data.
Next, we present a description and results of a Monte Carlo simulation we have performed to evaluate the EERD between the MLE and C-H classifiers for two multivariate normal configurations, ![]() ,
, ![]() , using various training-sample sizes, dimensions, features with block missing data, differences in means, values of correlation among variables, and missing-data proportions. For the simulations, we define p to be the total number of feature dimensions and r to be the number of missing features so that
, using various training-sample sizes, dimensions, features with block missing data, differences in means, values of correlation among variables, and missing-data proportions. For the simulations, we define p to be the total number of feature dimensions and r to be the number of missing features so that![]() . Also,
. Also, ![]() denotes the total training-sample size from population
denotes the total training-sample size from population![]() ,
, ![]() , and
, and
![]()
is the intraclass covariance matrix where ![]() denotes the common population correlation among the features in the intraclass covariance matrix and
denotes the common population correlation among the features in the intraclass covariance matrix and ![]() denotes a matrix of ones.
denotes a matrix of ones.
The simulation was performed in SAS 9.2 (SAS Institute In., Cary, NC, USA) using the RANDNORMAL command in PROC IML to generate 10,000 training-sample sets of size![]() ,
, ![]() , for each parameter configuration. Next, the MLE and C-H classifiers were computed, and their CERs were calculated for each training-sample set. Then, the differences between the CERs for the classifiers were averaged over the 10,000 CER differences for the two classifiers for each parameter configuration involving
, for each parameter configuration. Next, the MLE and C-H classifiers were computed, and their CERs were calculated for each training-sample set. Then, the differences between the CERs for the classifiers were averaged over the 10,000 CER differences for the two classifiers for each parameter configuration involving![]() , p, r,
, p, r, ![]() ,
, ![]() , and POMD for the r features with monotone missing data, where
, and POMD for the r features with monotone missing data, where![]() . Thus, the
. Thus, the ![]() for the C-H and MLES classifiers is
for the C-H and MLES classifiers is
![]()
where ![]() is defined in (12),
is defined in (12), ![]() is given in (21), K is the total number of simulated training-data sets, and j denotes the
is given in (21), K is the total number of simulated training-data sets, and j denotes the ![]() simulated training-data set, where
simulated training-data set, where![]() . We display the results of our two Monte Carlo simulations by graphing
. We display the results of our two Monte Carlo simulations by graphing ![]() against
against ![]() for various configurations of p, r,
for various configurations of p, r, ![]() ,
, ![]() ,
, ![]() , and POMD.
, and POMD.
The relationship between p and r was fixed at ![]() and
and![]() . We chose these specific values of p and r to evaluate
. We chose these specific values of p and r to evaluate ![]() when the proportion of variables with missing data were both small and large relative to p. The choice of r and
when the proportion of variables with missing data were both small and large relative to p. The choice of r and ![]() depended on the value of p, and we provide the values of p, r, and
depended on the value of p, and we provide the values of p, r, and ![]() used in the Monte Carlo simulation in Table 1.
used in the Monte Carlo simulation in Table 1.
Lastly, we chose ![]() such that
such that
![]() (22)
(22)
and ![]() such that
such that
![]() (23)
(23)
with ![]() and
and ![]() to assess
to assess ![]() for both small and large between-class separation. These values for
for both small and large between-class separation. These values for![]() ,
, ![]() , given in (22) and (23), were chosen because they are similar to the population means used in the simulation used in [1] . Furthermore, we contrasted (8) and (19) using POMD = 0.5, 0.8 for the r covariates with BMM data, and as in [1] , we chose
, given in (22) and (23), were chosen because they are similar to the population means used in the simulation used in [1] . Furthermore, we contrasted (8) and (19) using POMD = 0.5, 0.8 for the r covariates with BMM data, and as in [1] , we chose ![]() to avoid singularity of the estimated covariance matrices. The comparison criterion
to avoid singularity of the estimated covariance matrices. The comparison criterion ![]() is plotted against
is plotted against ![]() for various combinations of p, r,
for various combinations of p, r, ![]() ,
, ![]() , and POMD in Figure 1 and Figure 2,
, and POMD in Figure 1 and Figure 2,![]() . Although we simulated values for
. Although we simulated values for ![]() for
for![]() , we omitted the graphs for
, we omitted the graphs for ![]() because the graphs are similar to the plots for
because the graphs are similar to the plots for ![]() and
and![]() . The graphs for
. The graphs for ![]() can be obtained from the authors.
can be obtained from the authors.
Figure 1, Figure 2 illustrate that the ![]() is consistently positive for the values of p, r,
is consistently positive for the values of p, r, ![]() ,
, ![]() ,
, ![]() , and POMD examined here. Moreover, the figures indicate that the primary parameters that influence the dominance of the MLES classifier are
, and POMD examined here. Moreover, the figures indicate that the primary parameters that influence the dominance of the MLES classifier are ![]() and
and![]() ,
,![]() . For all feature dimensions considered here, the C-H and MLES classifiers were competitive for
. For all feature dimensions considered here, the C-H and MLES classifiers were competitive for![]() . More importantly, for
. More importantly, for![]() ,
, ![]() increased as
increased as ![]() increased for all p, r,
increased for all p, r, ![]() ,
, ![]() ,
, ![]() , and POMD considered here. The most noteworthy increase in the
, and POMD considered here. The most noteworthy increase in the ![]() was for
was for ![]() when
when![]() , where
, where ![]() increased by approximately 0.10. This increase occurred for all specified values of p, r,
increased by approximately 0.10. This increase occurred for all specified values of p, r, ![]() , and POMD, and, thus, supported the superiority of the MLES classifier in terms of EERD for these configurations. Additionally, we noted that
, and POMD, and, thus, supported the superiority of the MLES classifier in terms of EERD for these configurations. Additionally, we noted that ![]() when
when![]() ,
, ![]() , and other parameters are allowed to vary.
, and other parameters are allowed to vary.
The MLES classifier especially outperformed the C-H classifier when ![]() for
for![]() , as compared to when
, as compared to when![]() . The smaller values of
. The smaller values of ![]() for
for ![]() can be attributed to the fact that for a relatively large
can be attributed to the fact that for a relatively large
![]()
Table 1. Dimensions and sample sizes for the Monte Carlo simulation.
Mahalanobis distance when ![]() and
and![]() , the EERs for both classifiers are small, thus yielding a smaller
, the EERs for both classifiers are small, thus yielding a smaller![]() .
.
As we used a large number of simulation iterations, we obtained![]() , where
, where ![]() is
is
the grid of parameter vectors considered in the simulation. Thus, the relatively small estimated standard errors also support our claim that ![]() for
for ![]() for the parameter configurations considered here. As Figure 1, Figure 2 indicate, the contrasting values of p, r,
for the parameter configurations considered here. As Figure 1, Figure 2 indicate, the contrasting values of p, r, ![]() , and POMD contribute marginally, if at all, to
, and POMD contribute marginally, if at all, to![]() . Regardless of the combination of parameter values considered here, the MLES classifier dominates the C-H classifier in terms of
. Regardless of the combination of parameter values considered here, the MLES classifier dominates the C-H classifier in terms of![]() .
.
In summary, the simulation results indicated that the MLES classifier became increasingly superior to the C-H classifier as the correlation magnitude among the features with no missing data and the features with BMM data increased.
We remark that the standard errors for the ![]() in the [1] simulations are not sufficiently small enough to conclude a difference in the ERRs of the two competing classifiers. Hence, their claim that the C-H classifier outperforms the MLES classifier as the percent of missing observations increases is questionable.
in the [1] simulations are not sufficiently small enough to conclude a difference in the ERRs of the two competing classifiers. Hence, their claim that the C-H classifier outperforms the MLES classifier as the percent of missing observations increases is questionable.
We also performed a second Monte Carlo simulation whose results are not presented here. In this simulation, all fixed parameter values were equivalent to those of the first simulation except for ![]() in (23), where we chose 0.80 of the elements of
in (23), where we chose 0.80 of the elements of ![]() to be non-zero. Consequently, we obtained slightly different results from those of our first simulation. However, the MLES classifier still outperformed the C-H classifier for all para- meter configurations when
to be non-zero. Consequently, we obtained slightly different results from those of our first simulation. However, the MLES classifier still outperformed the C-H classifier for all para- meter configurations when![]() . These results suggest that for classification problems with equal intra-class covariance matrices the MLES classifier is superior to the C-H classifier when at least small correlation exists among the features with missing data and the features without missing data.
. These results suggest that for classification problems with equal intra-class covariance matrices the MLES classifier is superior to the C-H classifier when at least small correlation exists among the features with missing data and the features without missing data.
4. Two Real-Data Examples
4.1. Bootstrap Expected Error Rate Estimators for the C-H and MLE Classifiers
In this section, we compare the parametric bootstrap estimated ERRs of the C-H and MLES classifiers for two real-data sets each having two approximate multivariate normal populations with different population means and equal covariance matrices. First, we define the bootstrap ERR estimator for the C-H classifier,![]() . Let
. Let![]() ,
, ![]() , and
, and ![]() be the MLEs of
be the MLEs of![]() ,
, ![]() , and
, and![]() , respectively, defined in Theorem 1. Also, let
, respectively, defined in Theorem 1. Also, let![]() ,
, ![]() , and
, and ![]() be the bootstrap estimates of
be the bootstrap estimates of![]() ,
, ![]() , and
, and![]() , respectively, calculated using the parametric bootstrap training-sample data
, respectively, calculated using the parametric bootstrap training-sample data
![]() (24)
(24)
that is generated from![]() ,
,![]() . Then, conditioning on
. Then, conditioning on![]() ,
, ![]() , and
, and![]() , the bootstrap CERs for the C-H classifier are
, the bootstrap CERs for the C-H classifier are
![]()
for![]() ,
, ![]() , where
, where![]() ,
, ![]() , and
, and ![]() are similar in definition to
are similar in definition to![]() ,
, ![]() , and f in (7), (11), and (10), respectively, except that we use the bootstrap multivariate normal data in (24). Thus, assuming equal prior probabilities, the bootstrap CER for the C-H classifier is
, and f in (7), (11), and (10), respectively, except that we use the bootstrap multivariate normal data in (24). Thus, assuming equal prior probabilities, the bootstrap CER for the C-H classifier is
![]() (25)
(25)
Also, conditioning on![]() ,
, ![]() , and
, and![]() , the bootstrap CERs for the MLES classifier are
, the bootstrap CERs for the MLES classifier are
![]()
where ![]() is a complete unlabeled observation from
is a complete unlabeled observation from![]() ,
, ![]() is similar in definition to
is similar in definition to ![]() in (18), and
in (18), and![]() ,
,![]() . Given
. Given ![]() and
and![]() , we have
, we have
![]()
and given![]() ,
,
![]()
where
![]()
Thus, assuming equal a priori probabilities of belonging to![]() ,
, ![]() , for an unlabeled observation, we have
, for an unlabeled observation, we have
![]() (26)
(26)
Hence, the estimated parametric bootstrap EERD for the C-H and MLES classifiers is
![]() (27)
(27)
where j denotes the ![]() simulated training-data set for
simulated training-data set for![]() . We use (27) to compare the C-H and MLES classifiers for two real-data sets given in the following subsections.
. We use (27) to compare the C-H and MLES classifiers for two real-data sets given in the following subsections.
4.2. A Comparison of the C-H and MLE Classifiers for UTA Admissions Data
The first data set was supplied by the Admissions Office at the University of Texas at Arlington and imple- mented as an example in [1] . The two populations for the UTA data are the Success Group for the students who receive their master’s degrees (![]() ) and the Failure Group for students who do not complete their master’s degrees (
) and the Failure Group for students who do not complete their master’s degrees (![]() ). Each training sample is composed of ten foreign students and ten United States students. Each foreign student had 5 variables associated with him or her. The variables are X1 = undergraduate GPA, X2 = GRE verbal, X3 = GRE quantitative, X4 = GRE analytic, and X5 = TOEFL score. For each observation in both data sets, variables
). Each training sample is composed of ten foreign students and ten United States students. Each foreign student had 5 variables associated with him or her. The variables are X1 = undergraduate GPA, X2 = GRE verbal, X3 = GRE quantitative, X4 = GRE analytic, and X5 = TOEFL score. For each observation in both data sets, variables![]() ,
, ![]() ,
, ![]() , and
, and ![]() are complete; however,
are complete; however, ![]() contains monotone missing data. The UTA data set as seen in [1] can be seen in Table 2.
contains monotone missing data. The UTA data set as seen in [1] can be seen in Table 2.
Also, the common estimated correlation matrix for the UTA data is
![]() (28)
(28)
We remark that only one sample correlation coefficient in the last column of (28) has a magnitude exceeding 0.50, which reflects relatively low correlation among the four features without BMM data with the one feature having BMM data.
To estimate EERD for the C-H classifier (8) and the MLES classifier (19) for the UTA Admissions data, we determine![]() , given in (27), using 10,000 bootstrap simulation iterations with
, given in (27), using 10,000 bootstrap simulation iterations with![]() ,
, ![]() ,
, ![]() ,
,
and ![]() for
for![]() . Additionally, the parametric bootstrap multivariate normal distribution parameters, which are the MLEs for the multivariate normal population parameters given in Theorem 1, are
. Additionally, the parametric bootstrap multivariate normal distribution parameters, which are the MLEs for the multivariate normal population parameters given in Theorem 1, are
![]()
and
![]()
for the means of ![]() and
and![]() , respectively, with common covariance matrix
, respectively, with common covariance matrix
![]()
Subsequently, we obtained ![]() with
with![]() , which indicated that the C-H classifier yielded slightly better discriminatory performance compared to the MLES classifier for the UTA data. The fact that the C-H procedure slightly outperformed the MLES classifier for the UTA data set in terms of EERD is not surprising. In the UTA data set, relatively little correlation exists among many of the features, and the C-H classifier does not require or use information in the correlation between the features with no missing data and the features with missing data. However, the MLES classifier does require at least a moderate degree of correlation between some features with no missing data and the feature with missing data to yield a more effective supervised classifier than the C-H classifier.
, which indicated that the C-H classifier yielded slightly better discriminatory performance compared to the MLES classifier for the UTA data. The fact that the C-H procedure slightly outperformed the MLES classifier for the UTA data set in terms of EERD is not surprising. In the UTA data set, relatively little correlation exists among many of the features, and the C-H classifier does not require or use information in the correlation between the features with no missing data and the features with missing data. However, the MLES classifier does require at least a moderate degree of correlation between some features with no missing data and the feature with missing data to yield a more effective supervised classifier than the C-H classifier.
4.3. A Comparison of the C-H and MLE Classifiers on the Partial Iris Data
The second real-data set on which we compare the C-H and MLES classifiers is a subset of the well-known Iris data, which is one of the most popular data sets applied in pattern recognition literature and was first analyzed by R. A. Fisher (1936). The data used here is given in Table 3.
The University of Irvine Machine Learning Repository website provides the original data set, which contains 150 observations (50 in each class) with four variables: X1 = sepal length (cm), X2 = sepal width (cm), X3 = petal length (cm), and X4 = petal width (cm). This data set has three classes: Iris-setosa (![]() ), Iris-versicolor (
), Iris-versicolor (![]() ), and Iris-virginica (
), and Iris-virginica (![]() ). We have used a subset of the original Iris data set by taking only the first 20 obser- vations from
). We have used a subset of the original Iris data set by taking only the first 20 obser- vations from ![]() and
and ![]() and omitting the Iris-virginica group (
and omitting the Iris-virginica group (![]() ). We emphasize that the variables in the partial Iris data are much more highly correlated than the variables in the UTA data. The estimated correlation matrix is
). We emphasize that the variables in the partial Iris data are much more highly correlated than the variables in the UTA data. The estimated correlation matrix is
![]() (29)
(29)
In (29), all estimated correlation coefficients in the last column had a magnitude greater than 0.50, which reflects a moderate degree of correlation among the features![]() ,
, ![]() , and
, and![]() , and the feature
, and the feature![]() , which has BMM data.
, which has BMM data.
For the Iris subset data, which can be found in Table 3, we used 10,000 bootstrap iterations, ![]() ,
, ![]() ,
, ![]() ,
, ![]() , and
, and![]() , where
, where![]() , for calculating
, for calculating![]() . Hence, the overall proportion of missing observations for the Iris subset data is greater than that of the UTA data set. The bootstrap parameters corresponding to
. Hence, the overall proportion of missing observations for the Iris subset data is greater than that of the UTA data set. The bootstrap parameters corresponding to ![]() and
and ![]() are
are
![]()
and
![]()
respectively, with common covariance matrix
![]()
For the parametric bootstrap estimate for ![]() corresponding to the C-H and MLES classifiers applied to
corresponding to the C-H and MLES classifiers applied to
the subset of the Iris data set, we obtained ![]() with
with![]() , which indicated
, which indicated
that![]() . Consequently, because of the relatively large correlations among the variables with no missing data, namely,
. Consequently, because of the relatively large correlations among the variables with no missing data, namely, ![]() ,
, ![]() ,
, ![]() , and the variable with missing data,
, and the variable with missing data, ![]() , the MLES classifier convincingly outperforms the C-H classifier in terms of EERD. This evidence essentially contradicts the conclusion in [1] that the C-H classifier is superior to the MLES classifier when the proportion of observations with missing data is substantial, regardless of the covariance structure.
, the MLES classifier convincingly outperforms the C-H classifier in terms of EERD. This evidence essentially contradicts the conclusion in [1] that the C-H classifier is superior to the MLES classifier when the proportion of observations with missing data is substantial, regardless of the covariance structure.
5. Conclusions
In this paper, we have considered the problem of supervised classification using training data with identical BMM data patterns for two multivariate normal classes with unequal means and equal covariance matrices. In doing so, we have used a Monte Carlo simulation to demonstrate that for the various parameter configurations considered here, ![]() , not POMD, has the greatest impact on EERD. We have also concluded that the MLES classifier outperforms the C-H classifier for all considered parameter configurations involving intra-class covariance structures when
, not POMD, has the greatest impact on EERD. We have also concluded that the MLES classifier outperforms the C-H classifier for all considered parameter configurations involving intra-class covariance structures when ![]() and becomes an increasingly superior statistical classification procedure as
and becomes an increasingly superior statistical classification procedure as ![]() approaches 1. This conclusion essentially contradicts the simulation results of [1] .
approaches 1. This conclusion essentially contradicts the simulation results of [1] .
We also have compared the MLE and C-H classifiers on two real training-data sets using ![]() in (27). From the real data set in [1] , we have demonstrated that the C-H classifier can perform slightly better than the MLES classifier. Moreover, we have used a subset of the prominent Iris data set from [9] to illustrate that when the magnitude of the correlation among features without missing data and features with missing data is moderate to large, the MLES classifier is superior to the C-H classifier.
in (27). From the real data set in [1] , we have demonstrated that the C-H classifier can perform slightly better than the MLES classifier. Moreover, we have used a subset of the prominent Iris data set from [9] to illustrate that when the magnitude of the correlation among features without missing data and features with missing data is moderate to large, the MLES classifier is superior to the C-H classifier.