Expression of the Genes OsNRT1.1, OsNRT2.1, OsNRT2.2, and Kinetics of Nitrate Uptake in Genetically Contrasting Rice Varieties ()
1. Introduction
Rice is a staple food for over half the world’s people, especially in Asia, Africa and Latin America, being especially important for the sustenance of poor people [1] . However, its widespread cultivation in tropical soils with low availability of nitrogen (N) causes the need to develop varieties that are more efficient in the uptake and use of this element, to reduce the need for application of nitrogen fertilizers.
In studies of the nitrogen use efficiency, nitrate (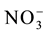 ) has great importance, because in aerated soils, it is the predominant form available to plants. The plant’s capacity to uptake and accumulate
) has great importance, because in aerated soils, it is the predominant form available to plants. The plant’s capacity to uptake and accumulate 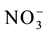 rapidly in the initial development phase can provide a greater stock of N for metabolism in the grain-filling phase [2] -[4] . Previous articles have reported that genetically contrasting rice varieties present variations in the uptake and use of
rapidly in the initial development phase can provide a greater stock of N for metabolism in the grain-filling phase [2] -[4] . Previous articles have reported that genetically contrasting rice varieties present variations in the uptake and use of , affecting the nitrogen use efficiency [4] [5] . Therefore, characterization of the kinetics parameters and expression levels of the
, affecting the nitrogen use efficiency [4] [5] . Therefore, characterization of the kinetics parameters and expression levels of the 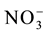 transporter genes is important to guide selection of varieties suitable for various growing conditions.
transporter genes is important to guide selection of varieties suitable for various growing conditions.
The objective of this study was to select, among local varieties traditionally grown in the Brazilian state of Maranhão, four genetically contrasting ones for determination of the N uptake kinetics parameters and analysis of the expression of the genes that encode transporter proteins of nitrate of low (OsNRT1.1) and high-affinity (OsNRT2.1 and OsNRT2.2).
2. Materials and Methods
2.1. Genotypes Selection by RAPD (Random Amplified Polymorphic DNA)
Seeds of 66 rice varieties (56 landraces and 10 improved), both Brazilian and imported (Electronic Supplemen- tary Material), were disinfested with sodium hypochlorite (2%) and germinated on gauze in distilled water. Five days after germination (DAG), the seedlings were collected and used to extract total genomic DNA [6] . For the polymerase chain reactions (PCRs), 26 primers were selected from those described by Araújo et al. [7] , Areias et al. [8] , Bhuyan et al. [9] and Rabbani et al. [10] .
After tests and amplification condition adjustments, seven primers (OPA 04, OPA 07, OPA 19, OPD 10, OPE 17, OPH 05, OPP 02) were chosen based on the number of polymorphic bands and band reproducibility profile. The PCRs were performed in 20 µL of material, containing 25 ng of genomic DNA, 2.3 mM of MgCl2, 0.2 mM of dNTP, 0.2 µM of each primer, one unit of Taq recombinant polymerase DNA (Invitrogen) and 2 µL of the enzyme’s buffer, with the volume completed by adding ultrapure water (Invitrogen), at temperatures according to Areias et al. [8] .
The PCR products were revealed by electrophoresis in 1.5% agarose gel, prepared using TBE 1/2 X, for two hours at 100 V and then stained in an ethidium bromide solution (10 µg∙mL−1). The gel images were analyzed using the GelCompar II software (Applied Maths) and the band profiles were used to generate a genetic similarity matrix using the coefficient of Jaccard [11] . Finally, a genetic similarity dendrogram was plotted using the unweighted pair group method with arithmetic mean algorithm (UPGMA).
2.2. Evaluation of Gene Expression of Nitrate Transporter
To analyze the expression of the nitrate transporter genes with low (OsNRT1.1) and high affinity (OsNRT2.1 and OsNRT2.2) and to determine the uptake kinetics parameters, four genetically contrasting rice varieties were se- lected (IAC-47, Bico Ganga, Arroz de Revenda and Manteiga) from the genetic similarity dendrogram obtained by Random Amplified Polymorphic DNA (RAPD) analysis (Figure 1). The seeds were disinfested and germinated as described previously. Fourteen days after sowing, the seedlings, measuring about 10 cm in height, were transferred to plastic pots (four seedlings each) containing 600 mL of the nutrient solution of Hoagland and Arnon [12] modified to 2 mmol∙L−1 of 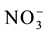 -N and other nutrients, with one-fourth the normal ionic strength (IS). Four days later, the nutrient solution was modified to 1/2 IS, while the
-N and other nutrients, with one-fourth the normal ionic strength (IS). Four days later, the nutrient solution was modified to 1/2 IS, while the 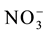 -N was maintained at 2 mmol∙L−1. The solution was renewed every three days thereafter. On 30 DAG, the plants were subjected to 72 h of total N deprivation, while maintaining the other nutrients at 1/2 ionic strength. After the deficiency period, the N supply was resupplied to one group of plants, with 0.2 mmol∙L−1 of nitrate, while the other group was kept in the solution without N. Root samples were collected 3, 6 and 9 h after resupply and stored at −80˚C for subsequent total RNA extraction.
-N was maintained at 2 mmol∙L−1. The solution was renewed every three days thereafter. On 30 DAG, the plants were subjected to 72 h of total N deprivation, while maintaining the other nutrients at 1/2 ionic strength. After the deficiency period, the N supply was resupplied to one group of plants, with 0.2 mmol∙L−1 of nitrate, while the other group was kept in the solution without N. Root samples were collected 3, 6 and 9 h after resupply and stored at −80˚C for subsequent total RNA extraction.
The total RNA was extracted according to Gao et al. [13] and the synthesis of the first cDNA strand was carried out from 0.5 µg of RNA, in a 96-well thermocycler (Eppendorf Mastercycler) using the High-Capacity cDNA Reverse Transcription Kit according to the manufacturer’s recommendations. The PCRs in real time were performed in duplicate in a Step One Plus Real Time PCR System using the SYBR® Green PCR Master Mix kit
![]()
Figure 1. Genetic similarity dendrogram based on the amplified DNA products of 66 rice varieties by RAPD, according to the Jaccard similarity coefficient.
(Applied Biosystems), according to the manufacturer’s recommendations. The sequences of the primers defined by Sperandio et al. [14] were used to amplify the nitrate transporters genes. Actin gene (OsAct) was used as an endogenous control [15] . The relative expressions were calculated according to Livak and Schmittgen [16] by relativization of the expression levels of the plants after resupply in function of the levels of those without resupply, so as to measure the levels of induction by the 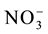 ion.
ion.
2.3. Nitrate Uptake Kinetics Assay
For the nitrate uptake kinetics assay, the Manteiga variety was excluded due to the absence of viable seeds. The germination of seeds of the other three varieties and the cultivation conditions in the growth chamber were the same as for the gene expression analysis. Immediately after N resupply and then each 30 minutes for the next 8 hours, 0.5-mL aliquots of nutrient solution were collected from each pot. At the end of this period, the volume of solution remaining in each pot was measured and the plants were collected and placed to dry at 60˚C in a forced-air chamber, after which the dried material was weighed.
The nitrate concentrations in the solution were determined according to Cataldo et al. [17] and were used to plot ion depletion curves and to determine the nitrate uptake kinetics parameters (Km and Vmax) through the mathematical graphing method proposed by Ruiz [18] , using the Cinética Win software developed by Ruiz and Fernandes Filho [19] . The values of Km and Vmax obtained for each replicate were submitted to analysis of variance and were used to estimate the 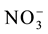 influx rates, using the Michaelis-Menten equation, as described by Epstein and Bloom [20] .
influx rates, using the Michaelis-Menten equation, as described by Epstein and Bloom [20] .
3. Results
3.1. Selection of Contrasting Genotypes by RAPD
The genetic similarity dendrogram obtained from the RAPD analysis presented two large groups (I and II) (Figure 1). Group I was highly heterogeneous regarding the origin of the varieties analyzed, including improved Brazilian and imported cultivars, new accessions to germplasma banks and a few from crops grown in the state of Maranhão.
The Piauí variety showed 88% similarity with IAC-600, a well-known variety called “black rice” developed by the Agronomic Institute of Campinas (São Paulo, Brazil). Although heterogeneous in its magnitude, Group I contained successive subdivisions that significantly separated the varieties included in it. Subgroup I.1a contained varieties with 100% similarity: Dourado Precoce and IAC-4440; Dobradinho and Zebu Branco. The improved variety IAC-47 was 95% similar to Dourado Precoce and IAC-4440. All the improved varieties were clustered in Group I. Most of the varieties with the name Lajeado from germplasma banks were also in Group I (Figure 1). It should be highlighted that Lageado Liso-220,029 and Lageado-220,006 had similarity greater than 80%, as also observed by Areias et al. [8] .
In turn, Group II presented the particular characteristic that it only contained varieties from crops grown in the county of Arari, Miranda do Norte, Penalva, Vitória do Mearim and Viana in Maranhão, suggesting a close genetic base for the varieties grown in that region (Figure 1).
Therefore, the similarity dendrogram allowed selecting four genetically contrasting rice varieties (IAC-47, Bico Ganga, Arroz de Revenda and Manteiga) for analysis of expression of the genes OsNRT1.1, OsNRT2.1 and OsNRT2.2 and determination of the nitrate uptake kinetics parameters.
3.2. The Expression Levels of Nitrate Transporter
Three hours after the resupply of nitrate, increases were observed of between 40 and 250 times in the expression of the OsNRT2.1 and OsNRT2.2 genes in relation to the plants maintained without nitrogen (Figure 2(a) and Figure 2(b)), while for OsNRT1.1 the expression levels were two times higher (Figure 2(c)). In general, OsNRT2.1 and OsNRT2.2 were strongly induced with the resupply following the suppression after 3 hours, while interestingly OsNRT1.1 presented higher expression in some varieties 6 and 9 hours later, suggesting a slower induction response (Figure 2(c)).
In the Arroz de Revenda and Manteiga varieties, the induction of expression of the OsNRT2.1 and OsNRT2.2 genes was greater with the resupply and was longer lasting (Figure 2(a) and Figure 2(b)), suggesting better adaptation to conditions of low soil nitrate levels.
3.3. Kinetic Parameters of the Nitrate Uptake
Due to the large differences regarding the induction of the NRTs, especially observed 3 hours after resupply, we also expected to see large differences in the nitrate uptake capacity between the varieties. However, no difference was observed between the varieties with respect to the curves of nitrate depletion in the nutrient solution and the uptake isotherms (Figure 3). There were no significant differences of Vmax between the treatments. The Km value of the Arroz de Revenda variety was higher than that of IAC-47, which had the lowest value of this parameter (Table 1).
4. Discussion
All the varieties studied presented higher expression of the OsNRT2.1 and OsNRT2.2 genes 3 hours after resupply. This result does not agree with that observed by Araki and Hasegawa [21] for the Nipponbare rice cultivar, for which these genes were induced by nitrate and exhibited peak expression about 3 hours after resupply. However, Hu et al. [22] , studying Arabidopsis thaliana, observed that the peak induction for AtNRT2.1 and AtNRT1.1 was reached only 30 minutes after resupply of nitrate, suggesting that the response time of the orthologues of NRTs can vary greatly from one species to another.
It is interesting to observe that the varieties with the highest levels of OsNRT2.1 and OsNRT2.2 transcripts during the study period were in nearly all cases those with the highest levels of OsNRT1.1 transcripts, principally at the intervals of 6 and 9 hours for the IAC-47 and Arroz de Revenda varieties. This relation can be due to the role of NRT1.1 as a constituent of the signaling mechanism for recognizing the presence of nitrate in the solution, as demonstrated in Arabidopsis [23] .
According to Girin et al. [24] , the expression of NRT2.1 is strongly suppressed by nitrate assimilation products. Therefore, the effects of induction by nitrate, free of suppression by feedback, explain the explosive
![]()
Figure 3. Nitrate depletion of the nutrient solution with 0.5 mmol∙L−1 of 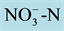 in three rice varieties. Each point represents the mean of three replicates. Vertical bars indicate the standard deviation from the mean of three replicates.
in three rice varieties. Each point represents the mean of three replicates. Vertical bars indicate the standard deviation from the mean of three replicates.
![]()
Table 1. Nitrate uptake kinetics parameters of three rice varieties after ressupply with 0.5 mmol∙L−1 of![]() .
.
Equal letters in the column do not differ between each other at 5% probability by the Tukey test. The values are the means of three replicates.
response of the OsNRT2 genes (induction of up to 250 times) in the first hours. According to Wirth et al. [25] , the OsNRT2.1 transporter is responsible for absorption of up to 75% of the available nitrate in Arabidopsis thaliana, while the huge differences regarding the induction response levels of the NRT2s, as observed for the varieties evaluated here, are not reflected in significant differences between the nitrate depletion curves (Figure 3), or between the Vmax values (Table 1) found in the second experiment. Regarding Km, the lower value obtained for the IAC-47 cultivar in relation to Arroz de Revenda (Table 1) suggests greater action of transporters with higher affinity. However, as demonstrated by Feng et al. [26] , it should be considered that to promote the absorption of nitrate, the OsNRT2.1/2.2 carriers require the participation of the protein NAR codified by the OsNAR2.1 gene, which was not evaluated in this study.
Other factors also might be involved in these varieties, balancing the capacity to uptake nitrate in detriment of differences in the levels of the transporters. Liu et al. [27] , studying the relative contribution of size and root activity for uptake of N between two contrasting strains of corn regarding N uptake efficiency, observed that the inefficient variety presented higher and more persistent expression levels of ZmNRT1.1, ZmNRT2.1, ZmNRT2.2 and ZmNAR2.1, while greater uptake efficiency was attributed to larger size and stronger growth response of the root system. Therefore, a more extensive root system in a variety with a lower NRT expression level could balance its capacity to absorb nutrients versus the other with high expression level. Nevertheless, we found no significant differences between the varieties regarding root dry mass. Therefore, the selection of genotypes for use in programs to improve N uptake efficiency also should consider other factors, such as the root system morphology.
Taken together, the data presented here demonstrated that expression of the high-affinity nitrate transporter genes of genetically contrasting rice varieties increased after resupply with this ion, with the highlight being the Arroz de Revenda variety, for which the expression induction was up to 250 times.
Acknowledgements
The authors are grateful to National Council for Scientific and Technological Development (CNPq), National Council for the Improvement of Higher Education (CAPES) and Carlos Chagas Filho Foundation for Research Support of Rio de Janeiro State (FAPERJ) for the scholarship and grants that supported this work. This work is part of the M.Sc. dissertation of the first author in Programa de Pós-Graduação em Agronomia-Ciência do Solo (CPGA-CS) at Universidade Federal Rural do Rio de Janeiro (UFRRJ).
Electronic Supplementary Material
List of rice varieties, including 56 landraces and 10 improved, used in the study with their accession number and origin, if known.
Varieties used in previous studies Souza et al. (1998), Ferraz Junior et al. (2001), Araújo et al. (2003), Areias et al. (2006) and Santos et al. (2007) or provided by ou fornecidas pelo CENARGEM (Embrapa), CNPAF (Embrapa) or collected in the Maranhão State (Brazil). The acession number of germplasm bank is presented when available. *Improved varieties.
NOTES
*Corresponding author.