1. Introduction
Recreational and decorative use of water can have important effects on health and well-being. Presently, in several cities, lakes and fountains are mainly used for these purposes. However, the contamination of urban lakes and fountains with fecal material can turn these water spots into potential hazards for public health. The presence of total coliforms, fecal coliforms and Escherichia coli is indicators of fecal pollution and commonly used to analyze the safety and quality of water supplies and recreational water [1,2].
E. coli belongs to the family of Enterobacteriaceae and is a common inhabitant of the gastrointestinal tract of humans and animals, and can be easily spread through water contaminated with fecal material. As a genetically diverse group, most strains of E. coli are harmless commensals, but others are capable of causing either intestinal or extra-intestinal (ExPEC isolates) disease, due to the existence of virulence genes in their genome or plasmids [3]. According to Clermont et al. [4], the analysis of virulence genes contributes to the classification of E. coli into phylogenetic groups. The most virulent strains belong to the phylogenetic group B2, with a minority of strains belonging to group D. Most of the commensal strains are included in group A and B1 [5].
Enterococci are also present in the normal gut of animals and humans, and have been used as a water quality indicator in many countries. Presently, the genus Enterococcus comprises 51 validly published species [6]. However, Enterococcus faecium and Enterococcus fecalis are the most prevalent species in animal feces and commonly used as indicators of fecal pollution [7,8].
In general, due to the difficulty associated with the direct isolation of pathogens from water samples, indicators of fecal pollution may denounce the presence of pathogenic microorganisms (such as salmonella) in water [9,10].
The presence of Salmonella spp. in water is an important parameter for the assessment of water safety, due to their high infective potential. Several characteristics of these bacteria, such as genetic (e.g. pathogenicity islands and virulence plasmids pSLT) or morphologic (e.g. fimbriae and flagella), facilitate their proliferation and adhesion to the eukaryotic cells of hosts, causing associated pathologies (manly gastroenteritis and typhoid fever) [11, 12].
Currently, all previous bacteria have been used in many studies in order to assess the burden of antimicrobial resistance present within a determined biome. Bacterial antimicrobial resistance is a current concern and there is a worrisome emergence and dissemination of resistance mechanisms due to the increasing use of antibiotics [13,14]. For example, in the case of some gram-negative bacteria (e.g. E. coli), antimicrobial resistance can be provided by the production of extended-spectrum β-lactamases (ESBL), which destroy antibiotics included in the class of betalactams [15,16].
Aquatic environments can provide the necessary conditions for the development and spreading of new or mutated microbial strains, mainly caused by the presence of low concentration of antibiotics [17]; the availability of diverse gene content is due to the possible contact with multiple transmission vectors of DNA [18-20] and a potential food source.
Previous studies have focused in water quality and safety of rivers [21] and coastal waters [22]. In this study, we assessed the presence of microorganisms, indicators of fecal contamination, in different recreational water spots, which are frequented by people or their household pets. Antimicrobial susceptibility of the isolates identified was also investigated. In addition, the detection of virulence genes was performed to assess the virulence of some E. coli isolates. The salmonella serotypification was also performed in order to predict their potential pathogenicity. The ultimate goal of this study is to alert for the risks and contribute to the improvement of public health conditions.
2. Materials and Methods
2.1. Sample Collection
Sampling was carried out between September 2011 and March 2012, in 30 places: 17 lakes and 13 fountains in Porto (Figure 1). Samples of 1000 ml of water were collected in sterile bottles, from two to four distinct points of the respective lake or fountain and immediately protected from sunlight and transported in an insulated container to the laboratory, where they were homogenized and subjected to a coarse filtration through sterile gauze, to remove major sediments. The following studies were
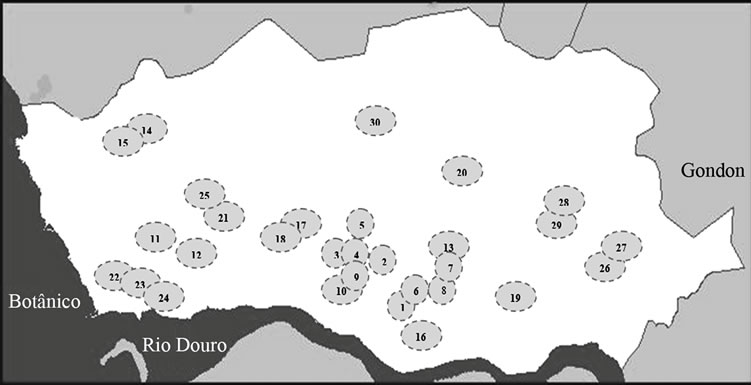
Figure 1. Map of the city of Porto, adapted from www. cmporto.pt. Each circle represents a sampling place (lakes-L and fountains-F): 1-Jardim da Cordoaria (L), 2-Jardim Abel Salazar (L), 3-Praça da Galiza (L), 4-Praça da Galiza (F), 5-Lago Rosália (L), 6-Fonte dos Leões (F), 7 and 8- Avenida dos Aliados (F), 9-Palácio de Cristal (L), 10-Palácio de Cristal (F), 11-Largo dos Arcos (L), 12-Arcos (F), 13-Largo da câmara municipal do Porto (F), 14 and 15-Parque da Cidade (L), 16-“Cubo” da Ribeira (F), 17-Jardim Botânico (L), 18-Jardim Botânico (F), 19-Campo 24 de Agosto (L), 20-Praça do Marquês (F), 21-Parque da Pasteleira (L), 22, 23 and 24- Jardim do passeio alegre (F), 25-Funda- ção Serralves (L), 26 and 27-Praça da Corujeira (L), 28 and 29-Parque de S. Roque (L) and 30-Jardim de Arca d’Água (L).
performed with the filtered sample.
2.2. Enumeration of Total Heterotrophs, E. coli and Enterococcus spp.
For E. coli, enterococci and total heterotrophs enumeration, duplicates of each water sample with volumes of 100 (for fountains), 50 (for lakes), 10, 1 and 0.1 ml were filtered through a 0.45 µm-pore-size membrane filters (Millipore Corporation, USA). The filters were placed on TBX Agar (Biokar Diagnostics, France), on Slanetz & Bartley Medium (SB) (Oxoid, United Kingdom) and on Plate Count Agar (Oxoid) for E. coli (blue-coloured colonies), enterococci (presuntive) and total heterotrophic bacteria enumeration, respectively. The plates were incubated at 37˚C for 24 h (E. coli) or 48 h (enterococci and heterotrophic bacteria). Enterococcus spp. colonies were confirmed in Kanamycin Aesculin Azide Agar (Liofilchem, Italy) (KAA), incubated at 44˚C for 4 hours (browncoloured colonies). Boxplot diagrams were designed with the CFU enumeration data of total heterotrophs, E. coli and Enterococcus spp., using the SPSS program version 19 for Windows (IBM Corporation, USA). The data were expressed as colony forming units (CFU) per 100 ml of water sample.
2.3. E. coli, Enterococcus spp. and Salmonella spp. Isolation and Antimicrobial Susceptibility Testing
For the isolation of salmonella, a volume of 100 ml was filtered through a 0.45 µm-pore-size membrane. A preliminary enrichment was carried out by placing the filter in 100 ml of Buffered Peptone Water (Oxoid). After incubation at 30˚C for 16 hours, the enrichment was performed transferring 1500 µl from the pre-enrichment to 15 ml of Selenite Cystine Enrichment Broth (Merck, USA) and 10 µl from the pre-enrichment to MRSV Agar (Biokar Diagnostics); both were incubated at 41.5˚C for 24 hours. Finally, the isolation of salmonella colonies was performed using the streak plate technique on XLD Agar (Biokar Diagnostics) and Hektoen Enteric Agar (Liofilchem) plates, incubated at 37˚C for 24 hours. Confirmation of the presence of salmonella was performed in TSI medium, API 20 E (BioMérieux, France) and antiserum polyvalent OMA (Bio-Rad, USA) through the observation of the formation of clots.
For the antimicrobial susceptibility testing and further PCR analysis, E. coli and enterococci isolates were obtained in the adequate selective media by the streak plate technique. Additionally, volumes of 50 to 100 ml of water samples were also filtered through a 0.45 µm-poresize membranes and then incubated on TBX agar plates supplemented with cefotaxime (2 µg/ml), ampicillin (8 µg/ml) or ciprofloxacin (4 µg/ml) (Sigma-Aldrich, France) and on SB supplemented with vancomycin (6 µg/ml), ampicillin (8 µg/ml) or ciprofloxacin (4 µg/ml), respectively. The plates were incubated at 37˚C during 24 - 48 hours.
A maximum of three colonies of E. coli and enterococci were selected from agar plates supplemented and nonsupplemented with antimicrobial drugs to test for antimicrobial susceptibility and then stored at −20˚C until further processing. Susceptibility to antimicrobial agents was tested using the disk diffusion method, according to the Clinical and Laboratory Standards Institute guidelines [23], using the following antibiotics (Oxoid) for E. coli and Salmonella spp.: cephalothin (CEF, 30 µg), cefoxitin (FOX, 30 µg), cefotaxime (CTX, 30 µg), ceftazidime (CAZ, 30 µg), amoxicillin/clavulanic acid (AMC, 30 µg), ampicillin (AMP, 10 µg), aztreonam (ATM, 30 µg), imipenem (IPM, 10 µg), gentamicin (GEN, 10 µg), kanamycin (KAN, 30 µg), tobramycin (TOB, 10 µg), amikacin (AMK, 30 µg), streptomycin (STR, 10 µg), nalidixic acid (NA, 30 µg), ciprofloxacin (CIP, 5 µg), chloramphenicol (CHL, 30 µg), tetracycline (TET, 30 µg), nitrofurantoin (NIT, 300 µg) and sulfamethoxazole/trimethoprim (SXT, 25 µg); and for Enterococcus spp.: ampicillin (AMP, 10 µg), gentamicin (GEN, 120 µg), ciprofloxacin (CIP, 5 µg), chloramphenicol (CHL, 30 µg), tetracycline (TET, 30 µg), nitrofurantoin (NIT, 300 µg), vancomycin (VAN, 30 µg), teicoplanin (TEC, 30 µg), erythromycin (ERY, 15 µg), azithromycin (AZM, 15 µg), rifampycin (RIF, 5 µg) and quinupristin-dalfopristin (Q/D, 15 µg).
The ESBL phenotype in E. coli culture was observed on plate according to the method of disk approximation test [15,24].
2.4. DNA Extraction
Genomic DNA was extracted by treatment with lysozyme (1 mg/ml; Sigma) and proteinase K (0.5 mg/ml; Sigma).
2.5. PCR Analysis and Salmonella spp. Serotypification
The screening of virulence genes and the determination of the phylogenetic groups of E. coli isolates were performed in 21 isolates from lakes (which displayed five or more antibiotic resistances) and 5 isolates from fountains (all the isolates from fountains that displayed antibiotic resistance). A group of 26 virulence genes reported in the literature to be associated with E. coli pathotypes were amplified by multiplex and uniplex PCR sets as previously described by Chapman et al. [25]. The phylogenetic groups of E. coli were determined with amplification of the genes chuA, yjaA and TSPE4.C2 [4].
Amplification of the genes related to the species-specific identification of E. fecalis, E. faecium, E. flavescens, E. durans, E. casseliflavus, E. gallinarum, E. avium, E. cecorum and E. hirae was performed as described by Jackson et al. [26]. This screening was performed in sixteen drug-resistant enterococci isolates.
PCR products were resolved by electrophoresis on a 1.5% agarose-Tris-Borate-EDTA gel containing ethidium bromide (10 mg/ml). A DNA molecular weight marker of 1kb (Plus DNA ladder, Fermentas, Lithuania) was used as a standard. The results were visualized by Gel Doc™ XR+ System with Image Lab™ Software (BioRad, USA).
The salmonella isolates were sent to LNIV (National Laboratory of Veterinary Investigation, Portugal) for serotypification.
3. Results
3.1. Enumeration of Bacterial Populations
All the sampled sites (17 lakes and 13 fountains) showed bacterial colonization. The average of CFU of heterotrophic bacteria per 100 ml of water sample detected for lakes and fountains was 1.32 × 105 and 4.23 × 104, respectively (Figure 2(A)). In addition, the CFU amount of E. coli was higher both in lakes (mean = 2.67 × 103 CFU/100 ml) and in fountains (mean = 3.52 × 102 CFU/100 ml) (Figure 2(B)), in comparison to enterococci CFU counts (mean = 5.60 × 102 CFU/100 ml and mean = 4.10 × 10 CFU/100 ml) (Figure 2(C)).
3.2. Antimicrobial Susceptibility and PCR Analysis
Resistance rates of E. coli and enterococci isolates from non-supplemented media are displayed in Tables 1 and 2, respectively. Interestingly, only resistance to cephalotin (9.1%) was found among the eleven E. coli isolates from fountains, whereas isolates obtained from lakes (n = 18) displayed resistance to tetracycline (44.4%), nalidixic acid (22.2%), cephalotin (16.6%) and similar resistance rates to ampicilin and streptomycin(27.8%), as well asto ciprofloxacin, chloramphenicol and kanamycin (16.7%). Resistance to cefotaxime, amikacin and trimethoprim/ sulfamethoxazol was observed only in 5.6% of E. coli isolated from lake samples.
In relation to the enterococci isolated from lakes (n = 35), a large number exhibited resistance to quinupristindalfopristin, tetracycline, rifampicin (>30%) and azitromicin (28.3%). Furthermore, resistance to erythromycin (17.1%), nitrofurantoin (14.3%), ciprof loxacin (11.4%), ampicillin (2.9%) and chloramphenicol (2.9%) was also observed. In contrast, among the 12 enterococci isolates

Figure 2. Enumeration of total heterotrophs (A); E. coli (B) and Enterococcus spp. (C) from lakes and fountains. The estimated means with the associated standard deviation are under the respective water spot name.
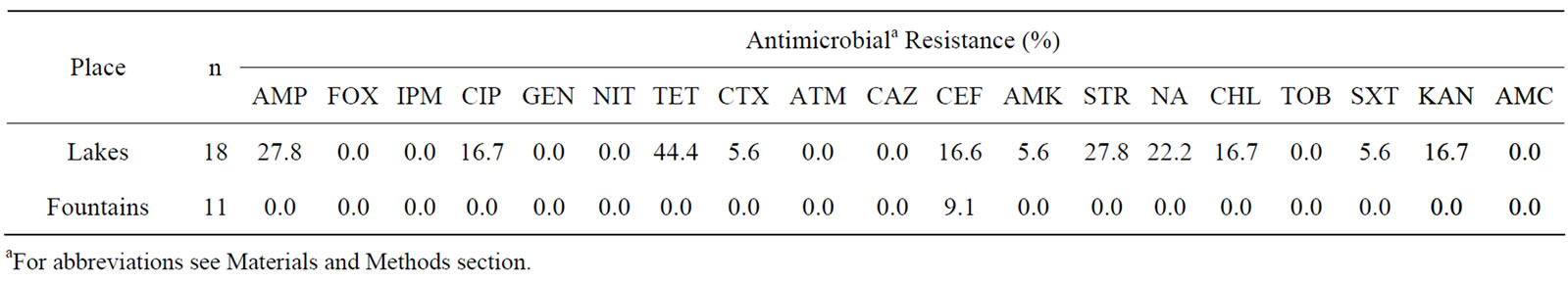
Table 1. Percentage of antibiotic resistance among 29 E. coli isolates obtained from non-supplemented media (18 isolated from lakes and 11 from fountains).
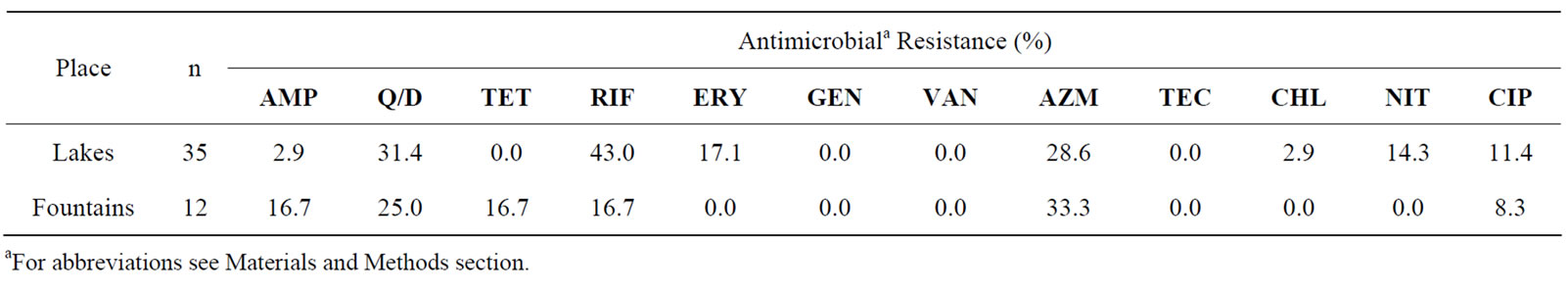
Table 2. Percentage of antibiotic resistance among 47 enterococci isolates obtained from non-supplemented media (35 isolated from lakes and 12 from fountains).
from fountains, 33.3% showed resistance to azitromicin, 25% to quinupristin-dalfopristin and 16.7% showed resistance to ampicillin, tetracycline and erythromycin. A smaller percentage of the isolates (8.3%) showed resistance to ciprofloxacin.
The antimicrobial resistance pattern of the isolates with a higher number of simultaneous resistances is shown in Table 3 for E. coli (n = 26) and in Table 4 for Enterococcus spp. (n = 16). Both bacteria groups showed diverse drug resistance profiles. In addition, seven E. coli

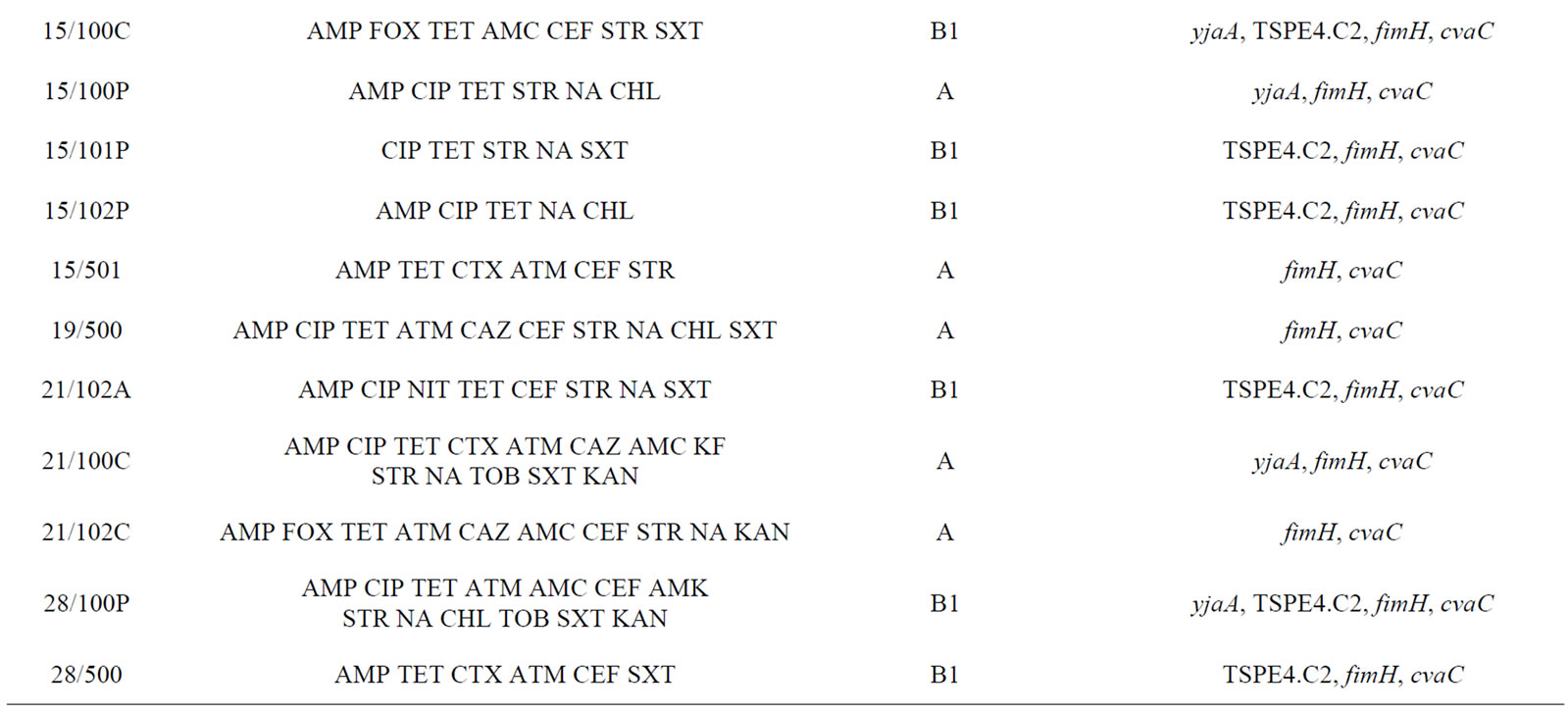
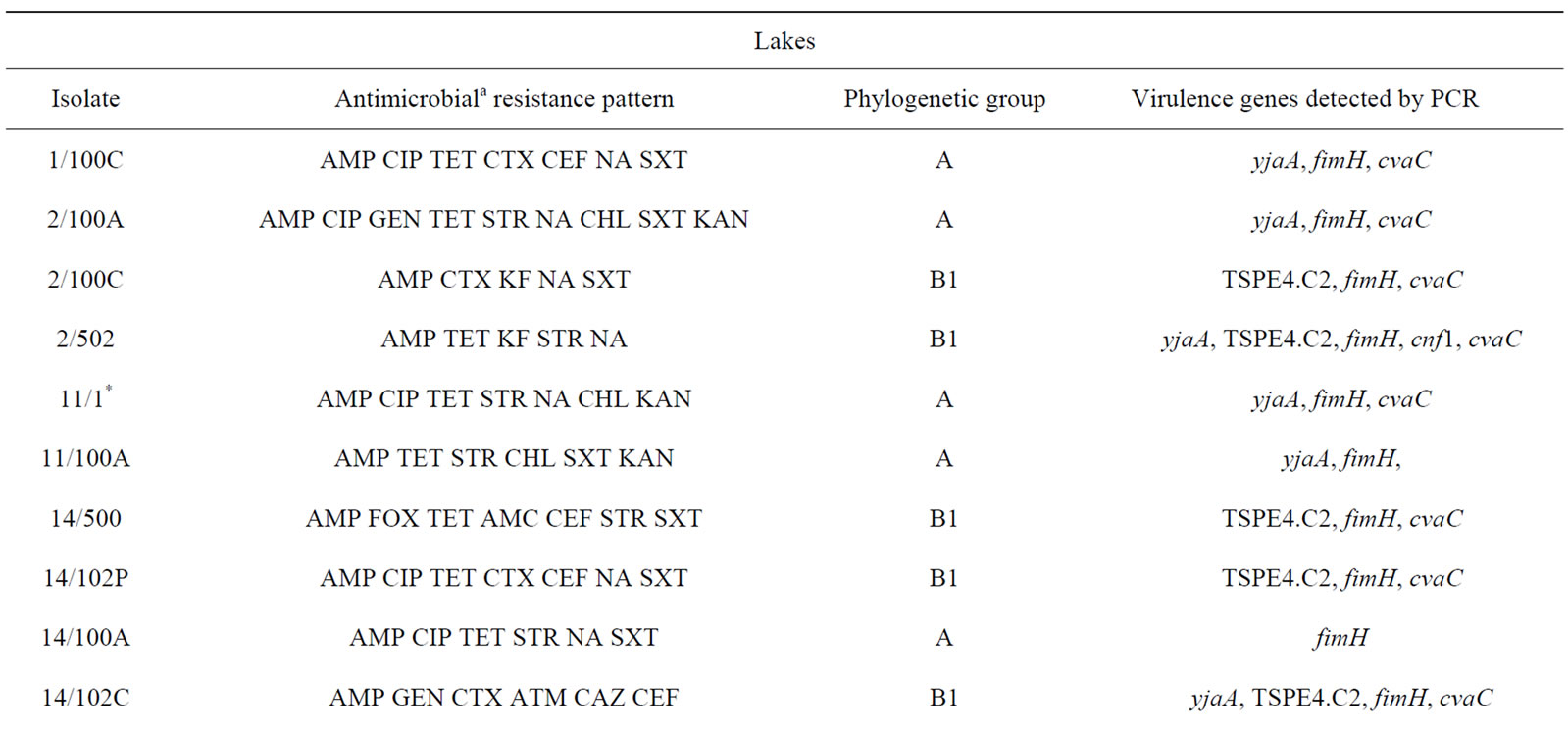
Table 3. Description of antimicrobial resistance profile, virulence genes and phylogenetic group of 26 E. coli isolates (21 from lakes and 5 from fountains).

Table 4. Description of antimicrobial resistance profile and enterococcal species of 16 isolates (9 from lakes and 7 from fountains).
isolates (2/100C, 2/502, 14/102P, 14/102C, 15/501, 21/100C and 28/500), exclusively from lake water samples, presented an ESBL phenotype.
Phylogenetic analysis of E. coli showed that the groups A and B1 were the most representative, with 14 and 11 isolates respectively. The phylogenetic group B1 appeared only in lake isolates and included five isolates with ESBL phenotype. In contrast, almost all E. coli isolates from fountains belonged to the phylogenetic group A, with the exception of isolate 23/1 which belonged to group B2. The virulence gene hlyA appeared exclusively in this isolate. The gene cnf1 was also amplified only in one E. coli isolate (2/502). On the other hand, fimH were found in all E. coli strains and only three isolates from lake water samples were negative for cvaC gene. The genes yjaA, TSPE4.C2 and chuA, which are used for determination of the phylogenetic group of E. coli, were identified in a total of 13, 12 and 1 isolate(s) respectively.
The drug-resistant enterococci isolates belong to five different species: E. faecium, E. fecalis, E. hirae and E. casseliflavus. Six out of nine enterococci isolates from lakes were identified as E. faecium and three as E. fecalis. Nevertheless, two isolat es from fountains were identified as E. fecalis and only one as E. faecium. Antibioticresistant isolates classified as E. hirae (one isolate) and E. casseliflavus (four isolates) were detected only in water samples from fountains.
Salmonella spp. was detected in two lake water samples. According to the salmonella serotypification performed by LNIV, both isolates corresponded to Salmonella II 42:b:e,n,x,z15. These isolates showed high susceptibility to all the antibiotics tested (data not shown).
4. Discussion
The importance of assessing the quality and safety of natural waters (as other natural habitats) have already been reported as being crucial to understand the phenomenon of antibiotic resistance in natural environments irrespective of the human use of antibiotics, and its potential relation with the development of antibiotic resistance in human pathogens [27,28].
Our quantitative results showed that heterotrophic bacteria, E. coli and Enterococcus spp. were more abundant in lakes than in fountains. This might be due to the fact that lakes have a greater environmental exposure (larger surface) and are exposed to the rainfall runoff, while fountains have less volume of water which is constantly renovated. Furthermore, the water of lakes is more turbid and stagnated, which decrease the bactericidal effects of UV radiation.
In this study, E. coli and enterococci were found to be ubiquitous among the water samples collected. Although tracing back the presence of these bacteria to their source is difficult; feces from birds (e.g. sparrows, seagulls and pigeons) [29,30], as well as other urban animals (such as dogs and cats) [31-33] are all possible sources. Actually, the lack of physical barriers allows the daily contact of pigeons, seagulls and/or ducks, as well as dogs, cats and/ or humans with these water spots, providing and “input and output” of the contaminant bacteria. In addition, droppings near water spots (on the ground or on trees for example) after dehydration may also be carried by the wind or pluvial waters into the lakes and fountains.
Regarding the antibiotic resistance profiles, many isolates displayed resistance to antibiotics commonly used in the treatment of bacterial infections, such as ampicilin, ciprofloaxacin and cefotaxime. However, only 7 out of 29 E. coli i solates and 10 out of 45 enterococci isolates from non-supplemented media showed a multidrug resistance profile (data not shown), whereas almost all supplemented media isolates (with antibiotic as a selective factor) showed multidrug resistance.
The majority of the multidrug-resistant enterococci studied were E. fecalis or E. faecium, but other species were also present at lower number such as E. casseliflavus and E. hirae. This finding is in agreement with previous studies that reported the prevalence of these strains in fecal contaminated environmental samples [34-36] and showed a particular ability of E. faecium and E. fecalis to acquire and maintain resistance traits [37,38].
Previous studies described a high prevalence of multidrug-resistant E. coli and Enterococcus spp. in wastewaters [35,39]. These habitats conjugate three important factors to the emergence and persistence of multidrug-resistant bacteria: i) a source of high microbial concentration and fecal bacteria; ii) a diverse pool of resistance genes; iii) the presence of sub-lethal antibiotic concentrations, which allow the development of antibiotic tolerance. In contrast, due to the absence of contact with the sewage, those factors should not be present in the water bodies sampled for this study. Thus, the results obtained suggest that the antimicrobial-resistant bacteria were added to the water spots and the expression of these resistances (e.g. ESBL) has been maintained. The presence of antibiotic-resistant bacteria in natural waters may contribute to the widespread dissemination of antimicrobial resistance, constituting a public health problem [13,28].
Although almost E. coli isolates were commensal it must be noted that these bacteria are able to interact and share their genomic content (e.g. virulence an antibiotic resistance genes) [20] with native microbiota in other places or hosts (such as in the human gut) [40]. Furthermore, the gene fimH was detected in all E. coli isolates. The expression of this gene is responsible for fimbriae adhesion to intestinal epithelium and colonization of the mucosa [41]. Previous studies indicated the high conservation of this gene in pathogenic E. coli from avian origin (APEC) (>99% homology), which is a subgroup of the extra-intestinal pathogenic E. coli pathotype (ExPEC) [42]. Additionally, the presence of the cvaC gene (which codifies a toxin) in the majority of the isolates tested (23 out of 26) also suggests the presence of E. coli from birds in these water spots. This gene is in a colicine V plasmid, which is commonly found in APEC and uropathogenic E. coli (UPEC) (another ExPEC group) and includes genes for antibiotic resistance [41,43]. This plasmid has also a high zoonotic potential, including for human microbiota [41,43]. Furthermore, UPEC strains are more likely to possess P pili, S pili, fimbrial adhesion, and toxins such as hemolysin and cytotoxic necrotizing factor 1 [3,44]. The last two features are related to the hlyA and cnf1 expression, respectively. In this study, only two isolates presented one of these genes: the isolate 2/502 which has the cnf1 gene and the isolate 23/1, which possesses the gene hlyA. The necrotizing factor 1 is a citotoxic exotoxin which is responsible for cell necrosis [45]. The hlyA induces proteolysis of host proteins, which modulates the epithelial cell functions and suppresses inflammatory responses (e.g. impairing the role of macrophages) [46]. The isolate 23/1 also belongs to the phylogenetic group B2, whose members are mainly pathogens [4].
PCR studies using combinations of three virulence genes [4] showed that almost all E. coli isolates were classified as belonging to the group A (chuA− yjaA− TSPE4.C2− or chuA− yjaA+ TSPE4.C2−) and B1 (chuA- yjaA− TSPE4.C2+), which together include almost all the commensal strains. In this way, the presence of some virulence genes is probably due to the high genome plasticity of E. coli strains, which is the main promoter for their rapid evolution, as reported in other studies [47,48].
According to standard conventions for salmonella serovar designation [49,50], the two salmonella isolates correspond to Salmonella enterica subsp. salamae serotype Uphill. Although this subspecies is not frequently associated to pathogenecity in humans, some reports from Centers for Disease Control and Prevention (CDC) and National Veterinary Services Laboratories from USA (NSVL) noticed this serotype as a clinical isolate from nonhuman sources (isolated from wild birds). Other studies showed a prevalence of Salmonella enterica salamae in reptiles and amphibians [51]. In addition, preliminary results from a study which we are conducting in the beaches of Porto showed a high prevalence of salmonella (77.3%) in seagull feces (n = 40) (unpublished data). Besides being hosts of Enterococcus spp. and E. coli, seagulls [52] and pigeons [53], as well as cats [54] and dogs [32] are usual vectors of salmonella. Similarly to our results, seagull sampled feces in another study [52] also showed high susceptibility to antibiotics, whereas the salmonella isolated from cats and dogs mentioned in the previous studies showed various profiles of antimicrobial resistance.
In our opinion, this fecal contamination could be prevented with the implementation of stronger environmental safety programs and specific legislation to ensure the water quality of these water supplies. Furthermore, a public awareness campaign (particularly for pet owners) and other preventive measures are also mandatory.
5. Conclusions
This study is the first report on fecal contamination associated with antibiotic resistance in lakes and fountains in Portugal. It is the clear evidence of fecal contamination in these water bodies. Lakes seem to be more affected by this kind of contamination. The well established presence of multidrug-resistant bacteria in these waters is also notorious, considering that there is not a direct source of antibiotics (the selective factor) in the places sampled.
Although almost E. coli isolates were commensal, some of them also have genes that give them the potential to cause opportunist infections in mammals (including humans) and birds, which come into contact with these contaminated waters daily. Moreover, the virulence genes detected also support the relation between bird feces and the contamination of these waters.
Even though these lakes and fountains are freely available to the population in general, their water safety and quality can be questionable. In fact, these water spots are not only a reservoir, but also a point for spreading fecal contaminants, including bacteria resistant to many of the antibiotics commonly used in human and veterinary medicine.
NOTES