Open Journal of Medical Microbiology
Vol. 3 No. 1 (2013) , Article ID: 28630 , 6 pages DOI:10.4236/ojmm.2013.31009
Analysis of Enterobacteriaceae Producing Broad-Spectrum Beta-Lactamases in the Intensive Care Unit Setting*
1Department of Microbiology, Faculty of Medicine and Dentistry, Palacký University Olomouc, Olomouc, Czech Republic
2University Hospital Olomouc, Olomouc, Czech Republic
3Department of Preventive Medicine, Faculty of Medicine and Dentistry, Palacký University Olomouc, Olomouc, Czech Republic
Email: #husickova.v@seznam.cz
Received March 30, 2012; revised May 9, 2012; accepted May 22, 2012
Keywords: Enterobacteriaceae; broad-spectrum beta-lactamases; intensive care setting
ABSTRACT
Strains of the Enterobacteriaceae family producing ESBL and AmpC broad-spectrum beta-lactamases that may survive in the hospital setting potentially cause infection in hospitalized patients due to contaminated objects or health care workers’ hands. Over a period of two months (November-December 2010), a single epidemiological study of microbial contamination of air, surfaces and health care workers (swabs from both nostrils and the right hand without a glove) was carried out at two intensive care units of the University Hospital Olomouc, Czech Republic. The bacteria were identified using standard microbiological methods. Phenotypic detection of ESBL and AmpC enzymes and basic genetic analysis of ESBLand AmpC-positive isolates was performed. The same approach was used to identify and analyze bacteria isolated from clinical samples of patients hospitalized at the above departments over the study period. From a total of 140 environmental samples collected over the study period, 21 isolates of the Enterobacteriaceae family were identified, with ESBL and AmpC production being detected in 4 and 7 isolates, respectively. Among patients’ clinical samples, 10 ESBLand 6 AmpC-positive isolates were detected. No similarity was found between environmental isolates and strains isolated from patients.
1. Introduction
Members of the Enterobacteriaceae family are feared nosocomial pathogens, especially in high-risk departments such as intensive care, neonatal and burn units. The risk stems mainly from an increasing prevalence of ESBLand AmpC-positive strains and the associated higher risk of failure of antibiotic therapy [1,2]. The growing number of Enterobacteriaceae spp. was also confirmed by data from the University Hospital Olomouc (UHO), Czech Republic. For instance, the percentage of Klebsiella pneumoniae isolates resistant to ceftazidime rose from 16% in 2005 to 40% in 2010 (unpublished data). The main mechanism of this resistance is production of broad-spectrum beta-lactamases, in particular ESBLs, that inhibit the effect of cephalosporin and penicillin antibiotics. Not uncommonly, isolates producing these enzymes are also resistant to other groups of antimicrobial agents such as fluoroquinolones, aminoglycosides, tetracyclines and co-trimoxazole [3,4]. In the UHO, fluroquinolone resistance of Klebsiella pneumoniae isolates increased from 12% in 2005 to 42% in 2010 (unpublished data). Besides Klebsiella pneumoniae strains, the common broad-spectrum beta-lactamase producers include Escherichia coli isolates and, less frequently, Enterobacter cloacae, Serratia marcescens, Citrobacter spp., Proteus spp. and others [5,6].
The prevalence of ESBL-positive strains also rises in “carrier” Enterobacteriaceae in the human gastrointestinal tract (GIT). Patients in the UHO were found to have 3% prevalence of ESBL-positive Enterobacteriaceae in the GIT in 2007 [7]. Carriage of these resistant bacteria is of particular importance to high-risk patients since they may cause endogenous nosocomial infections and serve as a reservoir for their further spread, including transferable genetic elements.
The potential sources of infection are patients and health care workers. However, suitable vehicles from the hospital environment may also play a role, in particular all wet areas where bacteria survive, e.g. sinks and drains. Most frequently, hand-touch sites are contaminated with Gram-negative bacteria, such as patient charts, tables, telephones and push plates [8]. There is clearly a dynamic relationship between the staff, patients and hospital environment, with bacterial strains being constantly transmitted [9].
With respect to the increasing bacterial resistance to antibiotics, measures have been introduced to lower the risk of nosocomial infections. These comprise antibiotic policy in hospitals including adequate application of antibiotics, proper hygienic disinfection of hands and the environment, isolation of the affected persons, search for contacts and, last but not least, continuous monitoring of resistant strains [10].
This prospective study aimed at determining the prevalence of Enterobacteriaceae producing ESBL and AmpC enzymes in two UHO intensive care units (ICUs) and performing their molecular biology analysis.
2. Material and Methods
Over a period of two months (November-December 2010), a single epidemiological study of microbial contamination of air, surfaces and health care workers (swabs from both nostrils and the right hand without a glove) was carried out in the UHO. The settings were the inpatient ward of the Department of Anesthesiology and Intensive Care Medicine (AIC; 10 beds) and the Department of Surgical Intensive Care Medicine (SIC; 12 beds).
Microbial contamination of air was determined using the MAS-100 microbial air monitoring system (Merck). At each department, samples were taken from a total of 10 sites. In all cases, 100 liters of indoor air were collected over a period of 1 minute.
Microbial contamination of surfaces was assessed using sterile swabs moistened with sterile saline. Samples were collected from a 10 × 10 cm area in two directions perpendicular to each other. In the case of sink and washbasin drains, samples were collected from the inner surfaces. At each department, 30 different surfaces were assessed (window boards, bed rails, infusion pump control panels, window blinds, sink drain, washbasin drain, rinse solution, tube stand).
Health care workers’ nasal mucosa was examined with a single swab of both nostrils. Samples were collected using sterile swabs moistened with sterile saline. Skin of the right hand without a glove was examined by swabbing the skin in places usually neglected during scrub-up, i.e. the thumb area and both the upper and lower sides of the interdigital spaces. Also in this case, samples were collected using sterile swabs moistened with sterile saline. At each department, 15 persons were examined.
Enterobacteriaceae were identified using standard microbiological methods including the ENTEROtest 16 (Erba-Lachema) and, in selected cases, the Phoenix automated system (BD Diagnostics) and the MALDI-TOF (Bruker Daltonics). The isolated bacteria were stored in a deep freezer at −80˚C using the ITEST Kryobanka B (ITEST plus). ESBL production was detected by the modified double-disk synergy test [11,12]. Phenotypic detection of AmpC beta-lactamases was carried out with the AmpC disk test with 3-aminophenylboronic acid [13, 14]. Genetic detection of beta-lactamases was performed using PCR and a set of specific primers for determining the presence of the blaTEM, blaSHV, blaCTX-M a blaAmpC genes [15]. SHV-products were digested with restriction endonuclease NheI, TEM-positive amplicons with the MseI, Sau3AI, MspI and HphI enzymes for identification of mutations extending the spectrum of activity [16,17]. To classify the present CTX-M beta-lactamases into individual groups, restriction analysis was performed using BseDI endonuclease [18]. To compare similar or identical isolates, pulsed-field gel electrophoresis (PFGE) of DNA fragments was used [19]. The resulting restriction profiles were compared using the GelComparII software (Applied Maths).
For comparison, Enterobacteriaceae were also isolated from clinical samples (tracheal secretions, bronchoalveolar lavage, sputum, blood, urine, pus, aspiration specimens, wound secretions) collected from patients hospitalized at the AIC and SIC departments during the same period. The strains were selected in such a manner that from each patient, only one strain for each species was included, which was isolated as the first one from the particular material.
3. Results
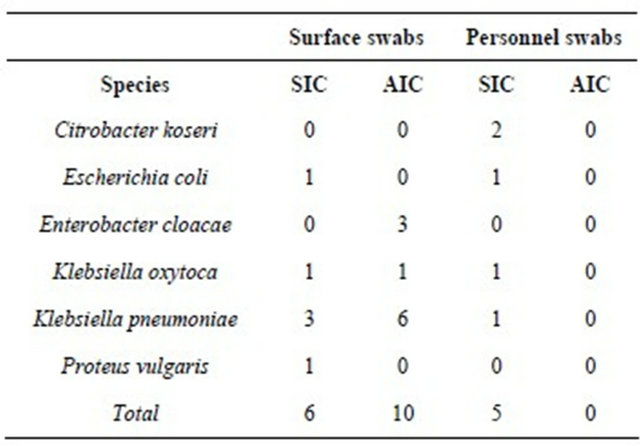
Over the study period, air samples for the assessment of bacterial contamination were collected from 20 sites. In addition, 60 swabs were taken from surfaces and 60 from health care workers (swabs from both nostrils and the right hand without a glove). From a total of 140 samples, 18 were found to contain 21 bacterial isolates of the Enterobacteriaceae family (Table 1).
Phenotypic analysis revealed ESBL production in 4 isolates; the AmpC disk test was positive in 7 cases. Environmental ESBLand AmpC-positive isolates including beta-lactamase types are summarized in Table 2.
ESBLand AmpC-positive Enterobacteriaceae isolated from clinical samples collected from patients at the aforementioned intensive care departments over the study period are listed in Table 3.
ESBLand AmpC-positive isolates from both the environment and clinical samples from hospitalized patients were subjected to genetic analysis aimed at the detection of individual beta-lactamase types. This revealed the presence of the blaTEM gene in 7 isolates, blaSHV gene in 10 isolates and blaCTX-M gene in 11 isolates. In 4 of the 10 genes carrying the blaSHV gene
Table 1. Numbers and species of isolates of the Enterobacteriaceae family detected in the intensive care unit setting (Department of Anesthesiology and Intensive Care Medicine-AIC; Department of Surgical Intensive Care Medicine-SIC).
restriction analysis detected mutations at position 238, associated with an extended spectrum of activity of the particular beta-lactamase. In the detected blaTEM genes, mutations extending the spectrum of activity were not confirmed by analyzing restriction fragment length. All the detected blaCTX-M genes were members of the CTX-M-like-1 group. Genes encoding AmpC beta-lactamases were observed in 13 isolates. Most frequently, a gene for the EBC type was identified. In only one case, a 405-bp amplicon was found, corresponding with the DHA type.
ESBLand AmpC-positive isolates were also subjected to restriction analysis of genome DNA followed by PFGE to determine similarity or identity. This comparison included strains of the same species isolated from the clinical samples collected from patients hospitalized at the respective centers in November to December 2010. Among 11 (6 environmental and 5 patient) Klebsiella pneumoniae isolates, PFGE identified four isolates with an identical restriction profile; the remaining 7 ones were genetically different. Identical isolates were also detected in Enterobacter cloacae (3/5). In both species, identical strains were only found among isolates from the hospital environment. In all cases, those were surface swabs. No similarity was found between environmental isolates and strains isolated from patients over the study period.
4. Discussion
The study documented the presence of ESBLand AmpC-positive Enterobacteriaceae in two intensive care units of the UHO. It is well known that in such types of departments, an increased use of antimicrobial agents is necessary, with selection pressure resulting in a high proportion of resistant strains. Of special importance are hygiene measures that are strictly adhered to in order to prevent the development of nosocomial infections and spread of resistant bacteria from the environment to patients and among them.
To emphasize the importance of adherence to hygiene practice and restriction of administration of third-generation cephalosporins, a study should be mentioned in which the above measures were necessitated by a high prevalence of ceftazidime-resistant Klebsiella pneumoniae strains at an intensive care unit. Such adherence
Table 2. ESBLand AmpC-positive Enterobacteriaceae detected in anesthesiology intensive care and surgical intensive care settings.
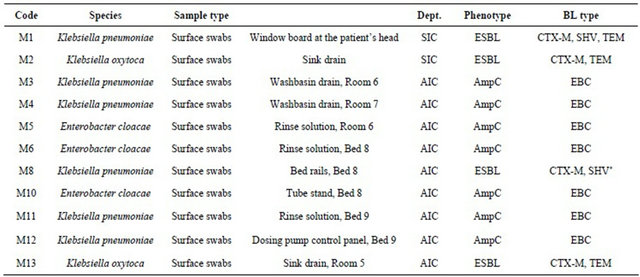
Legend: Dept.: Department; BL: Beta-lactamase; *Mutation extending the spectrum of activity detected by restriction analysis; AIC: Department of Anesthesiology and Intensive Care Medicine; SIC: Department of Surgical Intensive Care Medicine.
Table 3. ESBLand AmpC-positive patient isolates.
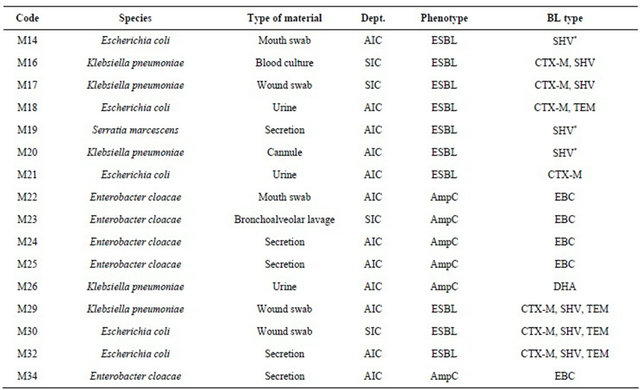
Legend: Dept.: Department; BL: Beta-lactamase; AIC: Department of Anesthesiology and Intensive Care Medicine; SIC: Department of Surgical Intensive Care Medicine.
proved to be highly effective, with a 4-fold increase in susceptibility to ceftazidime in Klebsiella pneumoniae isolates [10]. Although restrictive and barrier measures were only enforced in the ICU, there was a slight increase in susceptibility throughout the hospital [10].
Although the results of restricted use of antibiotics is positive it may not always be that significant. Toltzis et al. reduced the administration of ceftazidime at an ICU which resulted in a decrease, albeit insignificant, in ceftazidime-resistant Gram-negative bacteria. In the case of AmpC-positive bacteria, however, the prevalence dropped significantly from 68% to 46% [20].
Dancer et al. tried to measure the effect of enhanced cleaning of a surgical ward, with an additional cleaner. They found that such enhanced cleaning was associated with a 33% reduction in microbial contamination of hand-touch sites. Enhanced cleaning saved the hospital 30 to 70 thousand pounds [21].
Our study revealed the presence of ESBLand AmpCpositive Enterobacteriaceae in the ICU setting but no transmission of these strains to patients was noted. This suggests that hygiene precautions applied at such departments are adequate. However, the high rates of resistant Enterobacteriaceae in environmental swabs prove a marked resistance of these microorganisms to disinfection programs, a potential threat to patients. An international prospective study (Europe, Asia, Africa, North and South America, Australia) by Peterson provided alarming data: 31% (78 out of 253) of episodes of nosocomial bacteremia and 43% (30 out of 69) of episodes acquired at ICUs were caused by ESBL-producing bacteria. Previous administration of a beta-lactam antibiotic with an oxyimino group (cefuroxime, cefotaxime, ceftriaxone, ceftazidime and aztreonam) was associated with a higher risk of ESBL-positive bacteremia. In 7 out of 10 hospitals, interpatient transmission was also identified [22].
Our study did not detect ESBLand/or AmpC-positive Enterobacteriaceae in health care workers. According to Paterson et al., however, such a result does not rule out colonization of staff members with other multiresistant bacteria. In their study, no staff member carried ESBLproducing Escherichia coli but 23% of physicians and 32% of nurses had positive culture results that yielded methicillin-resistant Staphylococcus aureus or vancomycin-resistant Enterococcus species [23].
It may be concluded that there are effective measures for the control of multiresistant Enterobacteriaceae in the hospital setting, namely rational antibiotic therapy, strict adherence to hygiene precautions and education of both health professionals and the general public. However, another essential component of prevention is careful monitoring of the prevalence of resistant bacteria not only in patients’ clinical samples but also in the hospital environment, including genetic analysis of isolated strains.
REFERENCES
- C. M. Luna, P. Vujacich, M. S. Niederman, C. Vay, C. Gherardi, J. Matera and E. C. Jolly, “Impact of BAL Data on the Therapy and Outcome of Ventilator-Associated Pneumonia,” Chest, Vol. 111, No. 3, 1997, pp. 676-685. doi:10.1378/chest.111.3.676
- M. Tumbarello, M. Sanguinetti, E. Montuori, E. M. Trecarichi, B. Posteraro, B. Fiori, R. Citton, T. D’Inzeo, G. Fadda, R. Cauda and T. Spanu, “Predictors of Mortality in Patients with Bloodstream Infections Caused by Extended-Spectrum-ß-Lactamase-Producing Enterobacteriaceae: Importance of Inadequate Initial Antimicrobial Treatment,” Antimicrobial Agents and Chemotherapy, Vol. 51, No. 6, 2007, pp. 1987-1994. doi:10.1128/AAC.01509-06
- S. Müller, A. Oesterlein, M. Frosh, M. Abele-Horn and G. Valenza, “Characterization of Extended-Spectrum BetaLactamases and qnr Plasmid-Mediated Quinolone Resistance in German Isolates of Enterobacter Species,” Microbial Drug Resistance, Vol. 17, No. 1, 2011, pp. 99- 103. doi:10.1089/mdr.2010.0114
- C. H. Lee, J. W. Liu, C. C. Li, C. C. Chien, Y. F. Tang and L. H. Su, “Spread of ISCR1 Elements Containing blaDHA-1 and Multiple Antimicrobial Resistance Genes Leading to Increase of Flomoxef Resistance in ExtendedSpectrum-β-Lactamase-Producing Klebsiella pneumoniae,” Antimicrobial Agents and Chemotherapy, Vol. 55, No. 9, 2011, pp. 4058-4063. doi:10.1128/AAC.00259-11
- A. Sattar, F. Faqir, S. A. Abbasi, A. Fraz and Z. Hussain, “Changing Trends in Frequency of Extended-Spectrum Beta-Lactamase Producing Gram Negative Bacilli in Intensive Care Units of a Tertiary Care Hospital,” Pakistan Armed Forces Medical Journal, Vol. 59, No. 3, 2009, pp. 271-274.
- M. H. Sedlakova, V. Hanulik, M. Chroma, K. Hricova, M. Kolar, T. Latal, R. Schaumann and A. C. Rodloff, “Phenotypic Detection of Broad-Spectrum Beta-Lactamases in Microbiological Practice,” Medical Science Monitor, Vol. 17, No. 5, 2011, pp. 147-152. doi:10.12659/MSM.881761
- L. Cekanova, M. Kolar, M. Chroma, P. Sauer, M. Sedlácková and D. Koukalová, “Prevalence of ESBL-Positive Bacteria in the Community in the Czech Republic,” Medical Science Monitor, Vol. 15, No. 7, 2009, pp. 202- 206.
- S. J. Dancer, “What’s under Your Bed?! ESBLs: Infection Control in the Hospital Environment,” SCIEH Weekly Report, Vol. 38, No. 50, 2004.
- D. Pittet, B. Allegranzi, H. Sax, S. Dharan, C. L. PessoaSilva, L. Donaldson, J. M. Boyce, WHO Global Patient Safety Challenge and World Alliance for Patient Safety, “Evidence-Based Model for Hand Transmission during Patient Care and the Role of Improved Practices,” The Lancet Infectious Diseases, Vol. 6, No. 10, 2006, pp. 641- 652. doi:10.1016/S1473-3099(06)70600-4
- A. Noy, R. Orni-Wasserlauf, P. Sorkine and Y. SiegmanIgra, “Epidemiology of Ceftazidime-Resistant Klebsiella pneumoniae in a Large University Hospital in Tel Aviv,” The Israel Medical Association Journal, Vol. 2, No. 12, 2000, pp. 908-911.
- V. Jarlier, M. H. Nicolas, G. Fournier and A. Philippon, “Extended Broad-Spectrum b-Lactamases Conferring Transferable Resistance to Newer b-Lactam Agents in Enterobacteriaceae: Hospital Prevalence and Susceptibility Patterns,” Clinical Infectious Diseases, Vol. 10, No. 4, 1988, pp. 867-878. doi:10.1093/clinids/10.4.867
- M. Kolář, “Clinical Significance of Broad-Spectrum bLactamases and Experience with Their Identification in Microbiological Practice,” Klinická Mikrobiologie a Infekční Lékařství, Vol. 13, No. 5, 2007, pp. 195-205.
- T. Yagi, J. Wachino, H. Kurokawa, S. Suzuki, K. Yamane, Y. Doi, N. Shibata, H. Kato, K. Shibayama and Y. Arakawa, “Practical Methods Using Boronic Acid Compounds for Identification of Class C β-LactamaseProducing Klebsiella pneumoniae and Eschericha coli,” Journal of Clinical Microbiology, Vol. 43, No. 6, 2005, pp. 2551-2558. doi:10.1128/JCM.43.6.2551-2558.2005
- M. H. Sedlakova, V. Hanulik, M. Chroma, K. Hricova, M. Kolar, R. Schaumann and A. C. Rodloff, “The Potential of Phenotypic Methods for Detecting ESBLand AmpCType Broad-Spectrum Beta-Lactamases,” Clinical Microbiology and Infection, Vol. 16, No. 2, 2010, p. S152.
- M. Kolar, J. Bardon, M. Chroma, K. Hricova, T. Stosova, P. Sauer and D. Koukalova, “ESBL and AmpC BetaLactamase-Producing Enterobacteriaceae in Poultry in the Czech Republic,” Veterinary Medicine, Vol. 55, No. 3, 2010, pp. 119-124.
- A. Chanawong, F. H. M’Zali, J. Heritage, A. Lulitanond and P. M. Hawkey, “Characterisation of Extended-Spectrum β-Lactamases of the SHV Family Using a Combination of PCR-Single Strand Conformational Polymorphism (PCR-SSCP) and PCR-Restriction Fragment Length Polymorphism (PCR-RFLP),” FEMS Microbiology Letters, Vol. 184, No. 1, 2000, pp. 85-89.
- G. Arlet, G. Brami, D. Décrè, A. Flippo, O. Gaillot, P. H. Lagrange and A. Philippon, “Molecular Characterisation by PCR-Restriction Fragment Length Polymorphism of TEM β-Lactamases,” FEMS Microbiology Letters, Vol. 134, No. 2-3, 1995, pp. 203-208.
- M. Pimkin and M. Edelstein, “Improved PCR Detection and Subtyping of CTX-M β-Lactamase-Encoding Genes,” 14th European Congress of Clinical Microbiology and Infectious Diseases, Prague, 1-4 May 2004.
- Z. Y. Shi, P. Y. Liu, Y. J. Lau, Y. H. Lin and B. S. Hu, “Epidemiological Typing of Isolates from an Outbreak of Infection with Multidrug-Resistant Enterobacter cloacae by Repetitive Extragenic Palindromic Unit b1-Primed PCR and Pulsed-Field Gel Electrophoresis,” Journal of Clinical Microbiology, Vol. 34, No. 11, 1996, pp. 2784- 2790.
- P. Toltzis, T. Yamashita, L. Vilt, M. Green, A. Morrissey, S. Spinner-Block and J. Blumer, “Antibiotic Restriction Does Not Alter Endemic Colonization with Resistant Gram-Negative Rods in a Pediatric Intensive Care Unit,” Critical Care Medicine, Vol. 26, No. 11, 1998, pp. 1893- 1899. doi:10.1097/00003246-199811000-00035
- S. J. Dancer, L. F. White, J. Lamb, E. K. Girvan and C. Robertson, “Measuring the Effect of Enhanced Cleaning in a UK Hospital: A Prospective Cross-Over Study,” BMC Medicine, Vol. 7, 2009, p. 28. doi:10.1186/1741-7015-7-28
- D. L. Paterson, W. C. Ko, A. von Gottberg, S. Mohapatra, J. M. Casellas, H. Goossens, L. Mulazimoglu, G. Trenholme, K. P. Klugman, R. A. Bonomo, L. B. Rice, M. M. Wagener, J. G. McCormack and V. L. Yu, “International Prospective Study of Klebsiella pneumoniae Bacteraemia; Implications of Extended-Spectrum ß-Lactamase Production in Nosocomial Infections,” Annals of Internal Medicine, Vol. 140, No. 1, 2004, pp. 26-32.
- D. L. Paterson, N. Singh, J. D. Rihs, C. Squier, B. L. Rihs and R. R. Muder, “Control of an Outbreak of Infection Due to Extended-Spectrum Beta-Lactamase-Producing Escherichia coli in a Liver Transplantation Unit,” Clinical Infectious Diseases, Vol. 33, No. 1, 2001, pp. 126-128. doi:10.1086/320882
NOTES
*Supported by the following grant projects: LF_2012_006 and MSM 6198959223.
#Corresponding author.

