CellBio
Vol.3 No.2(2014), Article
ID:46735,10
pages
DOI:10.4236/cellbio.2014.32006
Intracellular Localization of ABC Transporter TAPL Differs between Transient and Stable Expression
Yu Motohashi1, Ayako Ohashi-Kobayashi2, Mayumi Nakanishi-Matsui3, Yasuyuki Fujimoto4, Masatomo Maeda4*
1Laboratory of Biochemistry and Molecular Biology, Graduate School of Pharmaceutical Sciences, Osaka University, Suita, Japan
2Department of Immunology, School of Pharmacy, Iwate Medical University, Shiwa-Gun, Japan
3Department of Biochemistry, School of Pharmacy, Iwate Medical University, Shiwa-Gun, Japan
4Department of Molecular Biology, School of Pharmacy, Iwate Medical University, Shiwa-Gun, Japan
Email: *mmaeda@iwate-med.ac.jp
Copyright © 2014 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 23 April 2014; revised 26 May 2014; accepted 2 June 2014
ABSTRACT
Transporter associated with antigen processing (TAP)-like (TAPL; ABCB9) is a half-type ABC transporter with sequence similarity to TAP1 and TAP2 that function in the ER membrane. To determine the cellular localization, TAPL and truncated forms of it were tagged with GFP at their carboxyl termini. Intracellular localization of these fusion proteins was compared between transient and stable expression in CHO-K1 cells. When they were expressed transiently, the fluorescence of the fusion proteins was detected on the intracellular membrane, mainly in the ER, and all the fusion proteins, i.e., TAPL(M1-A766)-GFP, TAPL(M1-S275)-GFP, TAPL(M1-K182)-GFP, TAPL(M1-R141)-GFP and TAPL(M1-G75), were co-localized with an ER marker, PDI. However, the fluorescence of all of them except for TAPL(M1-G75)-GFP and TAPL(M1-S275)-GFP overlapped with a lysosome marker, cathepsin D, upon stable expression. Lysosomal localization was similarly observed with TAPL(M1- A766)-DsRed, which was stably expressed. These results suggest that TAPL is sorted to the lysosomal membrane when expressed stably in CHO-K1 cells. Furthermore, the lysosomal targeting signal may comprise the N-terminal four transmembrane helices since the N-terminal two transmembrane helices may not be enough to function as such a signal.
Keywords:ABCB9, Lysosome, Membrane Localization, Stable Expression, TAPL, Transfection

1. Introduction
The ATP-binding cassette (ABC) transporters transport a variety of substrates from inorganic and organic ions to various xenobiotics and biomolecules across membranes using the energy of ATP hydrolysis [1] . Furthermore, mutations of the transporter genes cause congenital diseases such as cystic fibrosis, adrenoleukodystrophy and bare lymphocyte syndrome [2] .
TAPL is a half-type ABC transporter that forms a homo-dimer [3] [4] . The TAPL molecule is highly homologous to each subunit of TAP composed of a hetero-dimer of TAP1 and TAP2 [3] . Consistent with the amino acid sequence similarity of these three proteins, the gene organizations of TAPL, TAP1 and TAP2 are highly conserved [5] . Furthermore, both TAPL and TAP are peptide transporters, but the specificities of their transport substrates are different [6] [7] . The TAP transports antigen peptides into the lumen of the endoplasmic reticulum (ER) from the cytoplasm [8] . However, physiological function of TAPL has not been unveiled. Thus it is important to determine the cellular localization of TAPL in detail.
It is postulated that TAPL has ten transmembrane helices [9] , and the following cytoplasmic ABC domain is conserved in the ABC transporter family [1] . The downstream six transmembrane helices and the ABC domain of TAPL form a core domain essential for homo-dimerization, while its N-terminal four transmembrane helices do not participate in the dimer formation by TAPL [4] . When the fusion protein comprising TAPL with the green fluorescent protein (GFP) at its C-terminus was stably expressed in Chinese hamster ovary (CHO)-K1 cells, it was localized on the lysosomal membrane [4] [10] . It is further suggested that the N-terminal four transmembrane helices of TAPL comprise an unidentified lysosomal targeting signal [4] .
However, transiently expressed TAPL and carboxyl terminal deletion constructs of TAPL fused with GFP [TAPL(M1-A766)-GFP, TAPL(M1-S275)-GFP and TAPL(M1-G75)-GFP] were distributed mainly in the ER membranes of Cos-1 and HEK-293 cells [5] [9] . In this study, we further compared their intracellular localizations together with those of TAPL(M1-R141)-GFP and TAPL(M1-K182)-GFP when they are transiently and stably expressed in CHO-K1 cells. Interestingly, the amino-terminal two transmembrane helices of TAPL were localized in the ER membrane upon stable expression as well as transient expression. We also confirmed the lysosomal localization of the full-length TAPL molecule with a different tag, Discosoma sp. red fluorescent protein (DsRed), instead of GFP.
2. Materials and Methods
2.1. Expression Plasmids for Truncated TAPL-GFP Fusion Proteins
The expression plasmids for TAPL(M1-A766)-GFP, TAPL(M1-S275)-GFP, TAPL(M1-R141)-GFP and TAPL(M1-G75)- GFP were described in our previous papers [4] [9] . The expression plasmid for TAPL(M1-K182)-GFP was constructed as follows; a DNA fragment encoding M1-K182 was amplified by means of the polymerase chain reaction method using primers TM070 (5’-ccagatctaaccagcaggATGcgg-3’) and YM125 (5’-cgtcgacCTTggtgtaggagag-3’) carrying BglII and SalI sites (underlined) upstream of the initiation codon and downstream of the K182codon, respectively (bold capital letters) [preheating (94˚C, 5 min), followed by 30 cycles of denaturation (94˚C, 30 sec), annealing (64˚C, 30 sec), and extension (72˚C, 30 sec), and then post-incubation (72˚C, 7 min)]. The BglII-SalI fragment was inserted into the corresponding site of pEGFP-N1 (Clontech). An expression plasmid for TAPL(M1-A766)-DsRed was described previously [9] . The molecular biological methods used for DNA manipulations were based on the standard procedures [11] . DNA sequences were determined by means of the dideoxy chain-termination method [12] .
2.2. Cell Culture and Transfection
CHO-K1 cells were cultured in Ham’s F-12 medium (GIBCO) supplemented with 7% (v/v) fetal bovine serum (JRH Biosciences) [13] . Transient transfection was carried out as follows: on a Lab-tek II 4-well chamber slide (NalgenNunc), 5 × 103 cells were seeded in 1 ml of the medium and then cultured overnight. FuGENE6 (Promega) was used for transient transfection of plasmid DNA (1 μg), the plasmid solution being mixed with 98.5 μl of the medium (without antibiotics and serum) and 1.5 μl of FuGENE6. The mixture was processed for 30 min at room temperature and then poured into the culture medium. Cells were cultured further for 24 hr and subjected to analysis by immunofluorescence microscopy. Stable transformants were isolated in the presence of 0.1 mg/ml G418 (Nacalai) [13] .
2.3. Immunofluorescence Microscopy
The cells were fixed in methanol and then subjected to antibody-treatment as described previously [13] [14] . The antibodies [protein disulfide isomerase (PDI), cathepsin D and mannose-6-phosphate receptor (M6PR)] were diluted with 0.2% (w/v) gelatin in PBS, and then an aliquot (0.2 ml/well) was reacted at room temperature for 1 hr, followed by 5 min incubation with a gelatin solution (0.5 ml/well) three times. The second antibodies were similarly diluted (2000×) and used (1 ml/well). Cells were viewed under a microscope (Olympus IX70) equipped with an AQUACOSMOS U7501 (HAMAMATSU PHOTONICS). Cells were also incubated with either brefeldin A BODPY 558/568 or LysoTracker Red DND-99 (Molecular Probes) in the medium for 30 min [9] , and then monitored under a microscope as above.
2.4. Preparation of Soluble and Membrane Fractions
Cells (1.5 × 107) cultured for 2 days in a dish (Φ10 cm) were collected in 1 ml of ice-cold phosphate-buffered saline (PBS) [10 mM sodium phosphate buffer (pH 7.2), 137 mM NaCl, 3 mM KCl]. The cell pellet (12,000 × g, 5 min) was suspended in 0.5 ml 0.1M Tris-HCl (pH 7.5) containing protease inhibitors (3 μg/ml pepstatin A, 3 μg/ml leupeptin and 1 mM phenylmethylsulfonyl fluoride), and then sheered 70 times with a 26 G needle. Cell debris and nuclei were precipitated (750 × g, 5 min). The resulting supernatant was centrifuged (100,000 × g, 1 hr) (Beckman 50.3Ti rotor). The supernatant and precipitate were used as the soluble and membrane fractions, respectively. The membrane precipitate was suspended in 0.1 ml 0.1 M Tris-HCl (pH 7.5). Protein concentrations were determined with a BIO-RAD Protein Assay Kit [15] using bovine serum albumin (Fraction V, Sigma) as a standard.
2.5. SDS-Polyacrylamide-Gel Electrophoresis and Western Blotting
TAPL-GFP fusion proteins were analyzed by the published method [9] . Briefly protein samples were treated at 55˚C for 10 min and then subjected to sodium dodecyl sulfate (SDS)-polyacrylamide gel-electrophoresis [16] . Proteins were electro-blotted onto an ImmobilonTM-P filter (Millipore) [17] . The filter was reacted with antiGFP monoclonal peroxidase conjugated (6000× diluted, BD Biosciences), and then chemiluminescence was detected with an ECL Western Blotting Kit (GE Healthcare) using Scientific Imaging Film (KODAK).
2.6. Chemicals
Restriction enzymes and Ampli-Taq were obtained from New England Biolabs and Perkin Elmer, respectively. Agarose-LE Classic Type and Ligation Kit ver2 were purchased from TaKaRa. Oligonucleotides were purchased from Gene Design Inc. All other chemicals used were of the highest grade commercially available.
3. Results
3.1. Co-Localization of Transiently-Expressed TAPL-GFP Fusion Proteins with an ER Marker
CHO-K1 cells, transiently expressing TAPL derivatives tagged with GFP at their carboxyl termini (Figure 1), were fixed and processed for immunofluorescence detection using anti-PDI rabbit IgG and then Alexa 568- conjugated anti-rabbit mouse IgG. We compared the fluorescence of PDI (an ER marker) with that of TAPL derivatives, i.e., TAPL(M1-A766)-GFP, TAPL(M1-S275)-GFP, TAPL(M1-K182)-GFP, TAPL(M1-R141)-GFP and TAPL(M1-G75)-GFP. As shown in Figure 2(a), TAPL(M1-A766)-GFP was co-localized with the signal of PDI. The rest of the derivatives were distributed similarly to the signal of PDI (Figures 2(b)-(e)).
The GFP signals of TAPL and its derivatives were co-localized with brefeldin A BODPY 558/568 (an ER marker dye), but only partly with LysoTracker Red DND-99 (a lysosome marker dye) (Figure 3(A) and Figure 3(B), [a]-[e]). These results suggested that TAPL and its derivatives are mainly distributed in the ER membrane when expressed transiently in CHO-K1 cells, as demonstrated in Cos-1 cells and HEK-293 cells [9] . It should be noted that free GFP was distributed in the cytoplasm, but did not overlap with the ER marker signals of PDI and brefeldin A BODPY 558/568 (Figure 2(f), and Figure 3(A) [f] and Figure 3(B) [f], respectively).

Figure 1. Membrane topology of TAPL-GFP and its truncated derivatives. The membrane topology of human TAPL fused with GFP [TAPL(M1-A766)-GFP] is illustrated. Numerical values indicate the amino acid residue numbers at the ends of potential transmembrane helices. Helix numbers are also indicated by the underlined values in the center of transmembrane segments. The amino-terminus is indicated by “N”. The cytoplasmic catalytic domain is indicated by “ABC”. The truncated TAPL molecules with GFP [TAPL(M1-S275)-GFP, TAPL(M1-K182)-GFP, TAPL(M1-R141)-GFP and TAPL(M1-G75)-GFP] are also schematically shown.
3.2. Detection of TAPL-GFP Fusion Proteins in the Membrane Fraction
We examined whether the TAPL-GFP fusion proteins were localized in the membrane or not. A cell homogenate after transient expression was fractionated by ultracentrifugation (100,000 × g, 40 min), membrane and soluble fractions being prepared. Fusion proteins were probed by means of Western blotting with antibodies for GFP. As shown in Figure 4, all the fusion proteins were recovered in the membrane fraction (Figure 4(A), lanes 1 - 5), but not in the soluble fraction (cytoplasm) (Figure 4(B), lanes 1 - 5), whereas free GFP was only detected in the soluble fraction (Figure 4(A), lane 6 and Figure 4(B), lane 6). We concluded from these results that the GFP signals observed in Figure 2 and Figure 3 were due to the fusion proteins in the membrane.
Furthermore, the molecular masses on the gel (Figure 4(A), lanes 1 - 5 and Figure 4(B), lane 6) were closely similar to the sum of those of the GFP and TAPL moieties; the estimated values on the gel were 92, 54, 48, 43, (39 and 36), and 28 kDa, and the calculated values [determined with genetic information processing software GENTYXÒ MAC (Tokyo Japan)] were 114, 58, 48, 43, 36 and 29 kDa, respectively, for TAPL(M1-A766)-GFP, TAPL(M1-S275)-GFP, TAPL(M1-K182)-GFP, TAPL(M1-R141)-GFP, TAPL(M1-G75)-GFP and GFP, respectively. The doublet bands of the smallest construct, TAPL(M1-G75)-GFP, could be due to proteolytic cleavage or modification, since this fusion protein was distributed throughout the entire intracellular membrane and would thus have the chance to encounter various proteases or protein modification systems.
3.3. Comparison of GFP Signals of Stably Expressed TAPL-GFP Fusion Proteins with Those of Organelle Marker Proteins
Next we monitored the distribution of TAPL-GFP and its truncated versions stably expressed in the CHO-K1 cells. Previously, TAPL(M1-A766)-GFP and TAPL(M1-R141)-GFP were suggested to be localized to lysosomes, as determined by organelle staining with a fluorescent dye [4] . In this study, we detected the immunofluorescence of organelle marker proteins such as cathepsin D (a lysosome marker) and PDI (an ER marker) on microscopy.
As shown in Figure 5(a), TAPL(M1-A766)-GFP was co-localized with cathepsin D but not with PDI. Furthermore, TAPL(M1-R141)-GFP and TAPL(M1-K182)-GFP were sorted to lysosomes and were not retained in the ER (Figure 5(c), Figure 5(d)), suggesting that the amino-terminal four transmembrane helices (TM1 - TM4) were enough to target the TAPL molecule to the lysosomal membrane. The cytoplasmic loop between TM4 and
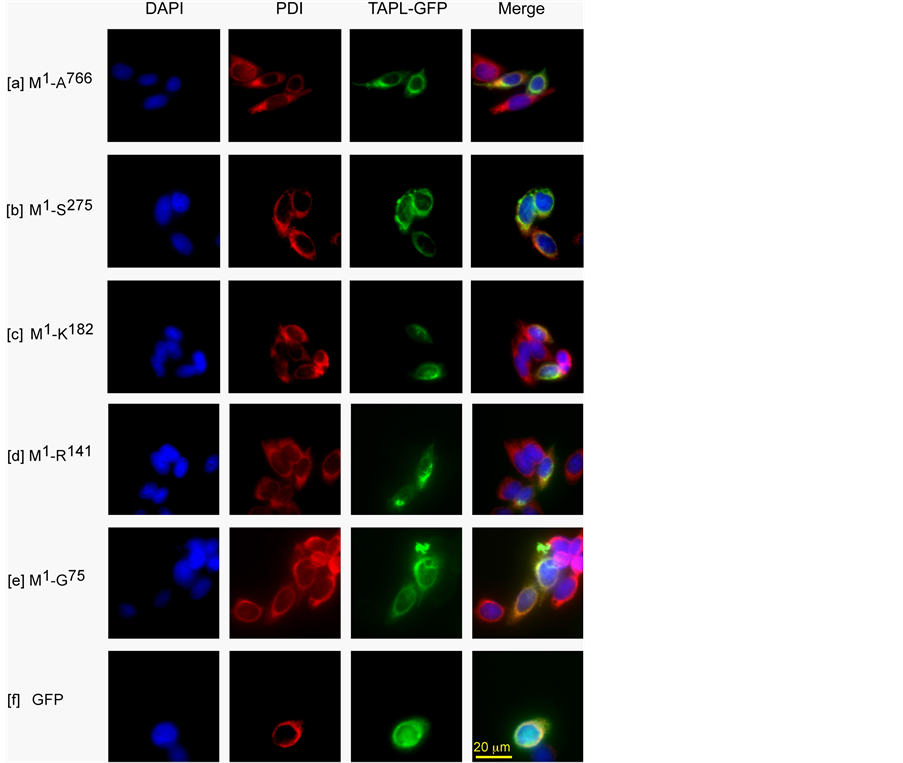
Figure 2. Cellular localization of transiently expressed TAPL-GFP and its truncated derivatives. CHO-K1 cells were transiently transfected with either the expression plasmid encoding full length human TAPL [TAPL(M1-A766)- GFP] or one of its truncated derivatives [TAPL(M1-S275)-GFP, TAPL(M1- K182)-GFP, TAPL(M1-R141)-GFP and TAPL(M1-G75)-GFP] tagged with GFP. The transfected cells were fixed and processed for immunofluorescence using anti-PDI rabbit immunoglobulin G (IgG) (1/100 dilution) and then Alexa 568-conjugated anti-rabbit mouse IgG (1/2000 dilution). Cells were imaged to localize an ER marker PDI (left center) and GFP (right center). The nuclei were stained with 4’,6-diamidino-2-phenylindole (DAPI) (left). All three figures were merged (right). From top to bottom; full length TAPL(M1-A766)- GFP [a], truncated derivatives with GFP carrying the M1-S275, M1-K182, M1-R141 and M1-G75 residues ([b]-[e], respectively), and GFP only [f]. Scales of all the figures are the same as [f] Merge (bar, 20 μm).
TM5 of TAPL(M1-K182)-GFP (Figure 1) had no effect on lysosomal targeting. Thus, the amino-terminal four transmembrane helices were enough for its targeting.
However, TAPL(M1-S275)-GFP was not co-localized with the signal of cathepsin D, as the GFP-signal was rather accumulated in cytoplasmic granules and ER (Figure 5(b)). An additional two transmembrane helices (TM5 and TM6) of truncated TAPL could be toxic and/or hinder the formation of the lysosomal signal even with their presence within the amino-terminal four transmembrane helices (TM1 - TM4). It was further demonstrated that the amino terminal two transmembrane helices (TM1 and TM2) were not enough to target TAPL to lysosomes, since TAPL(M1-G75)-GFP overlapped with the signal of PDI but not with that of cathepsin D (Figure 5(e)).

Figure 3. Co-localization of transiently-expressed TAPL-GFP and its derivatives with an ER marker. CHO-K1 cells transiently expressing TAPL-GFP or one of its truncated derivatives were incubated for 30 min with either brefeldin A BODPY 558/568 (ER marker) (A) or LysoTracker Red DND-99 (lysosome marker) (B). The cells were monitored under a microscope equipped for fluorescence emission detection (left; GFP signals, center; brefeldin A BODPY 558/568 (A) or LysoTracker Red DND-99 (B)). Merged figures are shown on the right. The overlapping of merged signals of GFP with ER and lysosome would be due to weak association of free GFP to membrane surface in the living cells, since free GFP was recovered in the soluble fraction as shown in Figure 4. Scales of all the figures in (A) and (B) are the same as [a] TAPL-GFP (A) and [a] TAPL-GFP (B), respectively, (bar, 20 μm).
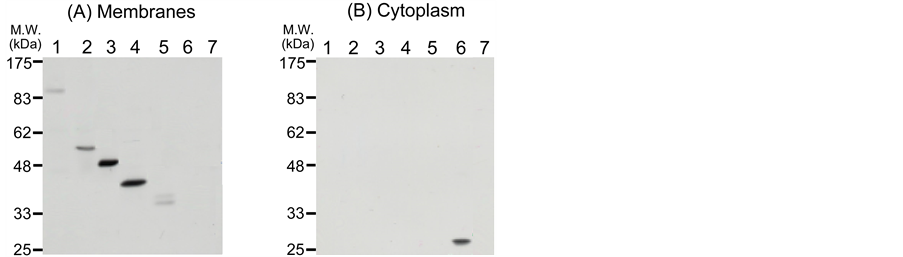
Figure 4. Detection of TAPL-GFP and its derivatives in membranes. Membrane (A) and soluble cytoplasmic (B) fractions (10 μg protein) prepared by the published method [25] were subjected to SDS-polyacrylamide gel-electrophoresis. GFP-fusion proteins were detected by means of Western blotting with anti-GFP antibodies. Lane 1, TAPL(M1-A766)-GFP; lane 2, TAPL(M1-S275)- GFP; lane 3, TAPL(M1-K182)-GFP; lane 4, TAPL(M1-R141)- GFP; lane 5, TAPL(M1-G75)-GFP; lane 6, GFP; lane 7, mock transfection with pME18S [10] .
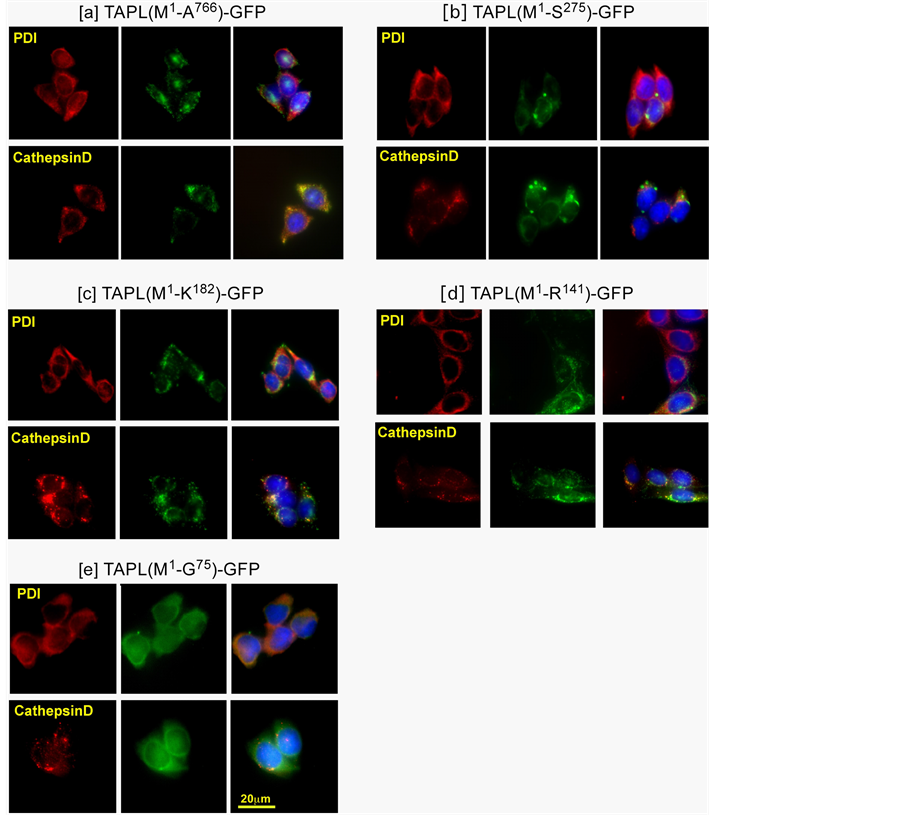
Figure 5. Cellular localization of stably-expressed TAPL-GFP and its truncated derivatives. TAPL-GFP or one of its truncated derivatives was stably expressed in CHO-K1 cells. The stable lines were fixed and processed for immunofluorescence using anti-PDI rabbit IgG or anti-cathepsin D rabbit IgG, and then Alexa 568-labeled anti-rabbit mouse IgG. Cells were imaged to localize either the marker proteins (left) or GFP signals (center). The nuclei were stained with DAPI and all three figures were merged (right). Scales of all the figures are the same as [e] Cathepsin D (merged) (bar, 20 μm).
3.4. Lysosomal Localization of TAPL Fused with DsRed
To further confirm the lysosomal localization of TAPL, we substituted the GFP moiety with a different fluorescent protein, DsRed. Stably expressed TAPL(M1-A766)-DsRed was co-localized with cathepsin D (a lysosomal marker), but not with PDI (an ER marker) or M6PR (a Golgi marker) (Figure 6). These results clearly indicated that the fluorescent protein moiety does not affect the lysosomal targeting of the TAPL molecule.
4. Discussion
In this study, we examined the membrane localization of TAPL under different expression conditions in detail. TAPL and carboxyl-terminal truncated forms of it were tagged with GFP. TAPL(M1-A766)-GFP, TAPL(M1- S275)-GFP, TAPL(M1-K182)-GFP, TAPL(M1-R141)-GFP and TAPL(M1-G75)-GFP were expressed transiently and stably in CHO-K1 cells, and the intracellular distribution of the fluorescence of GFP was compared with that of organelle markers on microscopy.
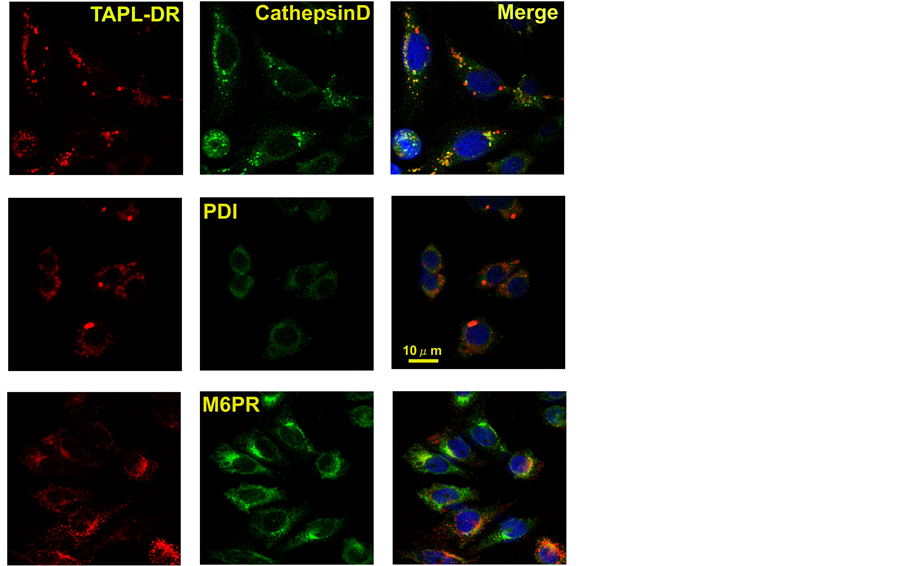
Figure 6. Lysosomal localization of TAPL-DsRed. TAPL-DsRed (TAPLDR) was stably expressed in CHO-K1 cells. Cells were treated (from top to bottom) with anti-cathepsin D, PDI or M6PR IgG, and then Alexa 488-labeled anti-rabbit mouse IgG. Cells were imaged to localize either the marker membrane proteins (center) or TAPL-DR (left). The nuclei were stained with DAPI. Merged figures are shown (right). Scales of all the figures are the same as PDI (merged) (bar, 10 μm).
The fluorescence of TAPL-GFP and its derivatives was co-localized with ER markers (PDI and brefeldin A BODPY 558/568), but only partly with lysosome markers (cathepsin D and LysoTracker Red DND-99) upon their transient expression in CHO-K1 cells. However, TAPL(M1-A766)-GFP, TAPL(M1-K182)-GFP and TAPL(M1- R141)-GFP as well as TAPL-DsRed expressed stably in CHO-K1 cells were co-localized with a lysosome marker protein, cathepsin D, rather than an endoplasmic reticulum marker, PDI. This localization is consistent with our previous findings [4] [10] , and that for a SKOV3 cell line derived from human ovaries [18] . The expression level would be too high upon transient expression, resulting in inhibition of the normal targeting pathway from the ER to lysosomes because various components for lysosomal targeting [19] , limiting amounts of which are present in the ER, could be saturated by the excess TAPL molecules. It is also possible that the increased amounts of TAPL derivatives tagged with GFP under transient expression could become targets of quality control system(s) of ER [20] and that the exit of TAPL molecules from ER to lysosomes through Golgi apparatus could be prohibited.
Stably expressed TAPL(M1-S275)-GFP and TAPL(M1-G75)-GFP were not localized to the lysosome membrane, while TAPL(M1-K182)-GFP and TAPL(M1-R141)-GFP were targeted to lysosomes. These results suggest that truncated TAPL with six transmembrane segments may be incapable of retaining a correct structure for membrane localization and thus may be accumulated in large granules, although the lysosomal targeting signal could comprise the N-terminal four transmembrane helices. There are three possibilities as to the lysosomal targeting signal; it is present in transmembrane helix 1 (TM1) and TM2, in TM3 and TM4, or TM1 - TM4. If the first possibility is the case, TAPL(M1-G75)-GFP could not attain the correct conformation without TM3 and/or TM4.
The sorting of transmembrane proteins involves recognition of short peptide signals in their cytoplasmic tails by cytoplasmic proteins [21] . Interestingly, the LLLWKN motif in the N-terminal cytoplasmic tail directs ABCA transporters (members of subfamily A of ABC transporters [1] ) to a post-Golgi vesicular sorting station [19] . Although TAPL, TAP1 and TAP2 are closely related ABCB subfamily members [5] , TAP (a heterodimer of TAP1 and TAP2) is localized to the ER and cis-Golgi [22] , while TAPL is sorted to lysosomes from the Golgi (this study and [4] [10] ). Interestingly, TAPL is 15 residues longer at the N-terminus than TAP1 and TAP2 [3] , the 3LWK5 sequence related to the LLLWKN motif being conserved in man and rodents [5] . It is of interest as to whether the N-terminal LWK sequence functions as a Golgi exit signal or not when TM3 and TM4 are assembled with TM1 and TM2 in TAPL.
In contrast, it is also claimed that transmembrane helices play a role in intracellular trafficking; the GxxxG motif and an arginine in the membrane-spanning domain are critical for interhelical interactions [23] . Such a GxxxG motif is present in TM2 of TAPL (63GATTG67) together with neighboring 58R and 70K [5] . Furthermore, related motif SxxxS [24] is also found in TM4 of TAPL (126SLGAS130). Thus, residues and motifs not only in cytoplasmic tails and loops but also in transmembrane segments should be focused on the search for lysosomal targeting signals for TAPL.
5. Conclusion
When a half-type ABC transporter, TAPL, and truncated forms of it were tagged with green fluorescent protein (GFP) at their carboxyl termini, the fusion proteins, i.e., TAPL(M1-A766)-GFP, TAPL(M1-S275)-GFP, TAPL(M1- K182)-GFP, TAPL(M1-R141)-GFP and TAPL(M1-G75)-GFP, were distributed on the intracellular membrane, mainly in the ER, upon transient expression in CHO-K1 cells. However, TAPL(M1-A766)-GFP, TAPL(M1-K182)- GFP, TAPL(M1-R141)-GFP and TAPL-DsRed were co-localized with a lysosome marker protein, cathepsin D, on stable expression. Stably expressed TAPL(M1-G75)-GFP was localized to the intracellular membrane. These results suggest that the lysosomal targeting signal comprised the amino-terminal four transmembrane helices, but amino-terminal two transmembrane helices were not enough to function as such a signal.
Acknowledgements
This research was supported in part by grants from MEXT for Strategic Medical Science Research Centers, 2010-2014 (The MIAST Project), and from JSPS (Grant-in-Aid for Scientific Research on Priority Areas B, 13142206) to M.M.
References
- Klein, I., Sarkadi, B. and Varadi, A. (1999) An Inventory of the Human ABC Proteins. Biochimica et Biophysica Acta, 1461, 237-262.
- Decottignies, A. and Goffeau, A. (1997) Complete Inventory of the Yeast ABC Proteins. Nature Genetics, 15, 137-145. http://dx.doi.org/10.1038/ng0297-137
- Yamaguchi, Y., Kasano, M., Terada, T., Sato, R. and Maeda, M. (1999) An ABC Transporter Homologous to TAP Proteins. FEBS Letters, 457, 231-236. http://dx.doi.org/10.1016/S0014-5793(99)01042-X
- Kamakura, A., Fujimoto, Y., Motohashi, Y., Ohashi, K., Ohahsi-Kobayashi, A. and Maeda, M. (2008) Functional Dissection of Transmembrane Domains of Human TAP-Like (ABCB9). Biochemical and Biophysical Research Communications, 377, 847-851. http://dx.doi.org/10.1016/j.bbrc.2008.10.078
- Kobayashi, A., Kasano, M., Maeda, T., Hori, S., Motojima, K., Suzuki, M., Fujiwara, T., Takahashi, E., Yabe, T., Tanaka, K., Kasahara, M., Yamaguchi, Y. and Maeda, M. (2000) A Half-Type ABC Transporter TAPL Is Highly Conserved between Rodent and Man, and the Human Gene Is Not Responsive to Interferon-Gamma in Contrast to TAP1 and TAP2. Journal of Biochemistry (Tokyo), 128, 711-718. http://dx.doi.org/10.1093/oxfordjournals.jbchem.a022805
- Koopman, J.O., Post, M., Neefjes, J.J., Hämmerling, G.J. and Momburg, F. (1996) Translocation of Long Peptides by Transporters Associated with Antigen Processing (TAP). European Journal of Immunology, 26, 1720-1728. http://dx.doi.org/10.1002/eji.1830260809
- Wolters, J.C., Abele, R. and Tampé, R. (2005) Selective and ATP-Dependent Translocation of Peptides by the Homodimeric ATP Binding Cassette Transporter TAP-Like (ABCB9). The Journal of Biological Chemistry, 280, 23631-23636. http://dx.doi.org/10.1074/jbc.M503231200
- Heemels, M.-T. and Ploegh, H. (1995) Generation, Translocation, and Presentation of MHC Class I-Restricted Peptides. Annual Review of Biochemistry, 64, 463-491. http://dx.doi.org/10.1146/annurev.bi.64.070195.002335
- Kobayashi, A., Maeda, T. and Maeda, M. (2004) Membrane Localization of Transporter Associated with Antigen Processing (TAP)-Like (ABCB9) Visualized in Vivo with a Fluorescence Protein-Fusion Technique. Biological and Pharmaceutical Bulletin, 27, 1916-1922. http://dx.doi.org/10.1248/bpb.27.1916
- Fujimoto, Y., Kamakura, A., Motohashi, Y., Ohashi-Kobayashi, A. and Maeda, M. (2011) Transporter Associated with Antigen Processing-Like (ABCB9) Stably Expressed in Chinese Hamster Ovary-K1 Cells Is Sorted to the Microdomain of Lysosomal Membranes. Biological and Pharmaceutical Bulletin, 34, 36-40. http://dx.doi.org/10.1248/bpb.34.36
- Sambrook, J., Fritsch, E.F. and Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual. 2nd Edition, Cold Spring Harbor Laboratory, Cold Spring Harbor.
- Sanger, F., Coulson, A.R., Barrell, B.G., Smith, A.J.H. and Roe, B.A. (1980) Cloning in Single-Stranded Bacteriophage as an Aid to Rapid DNA Sequencing. Journal of Molecular Biology, 143, 161-178. http://dx.doi.org/10.1016/0022-2836(80)90196-5
- Arimochi, J., Kobayashi, A. and Maeda, M. (2005) Stable Expression and Visualization of Mat-8 (FXYD-3) Tagged with a Fluorescent Protein in CHO-K1 Cells. Biotechnology Letters, 27, 1017-1024. http://dx.doi.org/10.1007/s10529-005-7870-4
- Arimochi, J., Ohashi-Kobayashi, A. and Maeda, M. (2007) Interaction of Mat-8 (FXYD-3) with Na+/K+-ATPase in Colorectal Cancer Cells. Biological and Pharmaceutical Bulletin, 30, 648-654. http://dx.doi.org/10.1248/bpb.30.648
- Bradford, M.M. (1976) A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Analytical Biochemistry, 72, 248-254. http://dx.doi.org/10.1016/0003-2697(76)90527-3
- Laemmli, U.K. (1970) Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature, 227, 680-685. http://dx.doi.org/10.1038/227680a0
- Towbin, H., Staehelin, T. and Gordon, J. (1979) Electrophoretic Transfer of Proteins from Polyacrylamide Gels to Nitrocellulose Sheets: Procedure and Some Applications. Proceedings of the National Academy of Sciences USA, 76, 4350-4354. http://dx.doi.org/10.1073/pnas.76.9.4350
- Zhang, F., Zhang, W., Liu, L., Fisher, C.L., Hui, D., Childs, S., Dorovini-Zis, K. and Ling, V. (2000) Characterization of ABCB9, an ATP Binding Cassette Protein Associated with Lysosomes. The Journal of Biological Chemistry, 275, 23287-23294. http://dx.doi.org/10.1074/jbc.M001819200
- Beers, M.F., Hawkins, A., Shuman, H., Zhao, M., Newitt, J.L., Maguire, J.A., Ding, W. and Mulugeta, S. (2011) A Novel Conserved Targeting Motif Found in ABCA Transporters Mediates Trafficking to Early Post-Golgi Compartments. Journal of Lipid Research, 52, 1471-1482. http://dx.doi.org/10.1194/jlr.M013284
- Sitia, R. and Braakman, I. (2003) Quality Control in the Endoplasmic Reticulum Protein Factory. Nature, 426, 891-899. http://dx.doi.org/10.1038/nature02262
- Kirchhausen, T. (2002) Single-Handed Recognition of a Sorting Traffic Motif by the GGA Proteins. Nature Structural Biology, 9, 241-244. http://dx.doi.org/10.1038/nsb0402-241
- Abele, R. and Tampé, R. (2004) The ABCs of Immunology: Structure and Function of TAP, the Transporter Associated with Antigen Processing. Physiology, 19, 216-224. http://dx.doi.org/10.1152/physiol.00002.2004
- Miyauch, K., Curran, A.R., Long, Y., Kondo, N., Iwamoto, A., Engelman, D.M. and Matsuda, Z. (2010) The Membrane-Spanning Domain of gp41 Plays a Critical Role in Intracellular Trafficking of the HIV Envelope Protein. Retrovirology, 7, 95. http://dx.doi.org/10.1186/1742-4690-7-95
- Curran, A.R. and Engelman, D.M. (2003) Sequence Motifs, Polar Interactions and Conformational Changes in Helical Membrane Proteins. Current Opinion in Structural Biology, 13, 412-417. http://dx.doi.org/10.1016/S0959-440X(03)00102-7
- Maeda, M., Nishijima, M., Akamatsu, Y. and Sakakibara, Y. (1985) Alteration in the Characters of CDP-Choline-synthetase and Phospholipid-Choline Exchange Enzyme upon Choline Starvation in Chinese Hamster Ovary Cells. The Journal of Biological Chemistry, 260, 5925-5930.
NOTES

*Corresponding author.

