Food and Nutrition Sciences
Vol.4 No.6(2013), Article ID:33051,8 pages DOI:10.4236/fns.2013.46089
Isolation and Characterization of Enterococci Bacteriocinic Strains from Tunisian Fish Viscera
![]()
1U12-ES03, Department of Biochemistry, Faculty of Medicine Ibn El Jazzar, University of Sousse, Sousse, Tunisia; 2Laboratoire Universitaire de Biodiversité et d’Ecologie Microbienne (EA3882), IFR148 ScInBioS, Université de Brest, Université Européenne de Bretagne, IUT de Quimper, 6 Rue de l’Université, Quimper Cedex, France.
Email: *taoufik_ghrairi@yahoo.fr
Copyright © 2013 Migaw Sarra et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received July 9th, 2012; revised March 5th, 2013; accepted March 13th, 2013
Keywords: Bacteriocins; Fish Viscera; Enterococcus; Listeria; Carnobacterium; Probiotic
ABSTRACT
A collection of lactic acid bacteria isolates from fish viscera was studied and investigated regarding to their functional properties and safety aspects. From these, three isolates GM1, GM2 and GM3 were identified as Enterococcus feacium species using molecular methods. Partial Amplified rDNA Restriction Analysis (partial ARDRA) with restriction enzyme HaeIII separated these isolates into distinctive group which suggest genotypic variability within enterococci strains isolated from fish viscera. The three strains GM1, GM2 and GM3 exhibited antimicrobial activity. Indeed strains have been shown to produce bacteriocins with inhibitory effect against food spoilage bacteria and pathogenic fish including Carnobacterium maltaromaticum. The molecular mass of bacteriocin, as calculated by tricine-SDS-PAGE, was found to be 4.5 kDa. All isolates were tested positive upon PCR amplification of enterocin A structural gene. Investigations of antibiotic resistance show that the isolates were mostly sensitive to several antibiotics (ampicillin, penicillin, tetracycline, gentamycine) and resistance to rifampicin. All isolates grow in esculin azide agar as a selective medium for enumeration of probiotic enterococci. This study suggests that our strains can be employed as probiotic or to improve the safety of food products.
1. Introduction
Enterococci belongs to the group of lactic acid bacteria (LAB) and are widely distributed in nature. The genera comprise more than 30 species, but Enterococcus faecium and Enterococcus faecalis are the most prevalent species in foods [1-4]. Some enterococci are bacteriocinogenic and capable to inhibit the growth of certain pathogens and spoilage microorganisms, presenting a great potential in food preservation [2,5].
It has been established that the intestinal microflora of fish is complex and that several species of LAB are part of the natural intestinal microflora of healthy fish [6]. Enterococci have been isolated from the intestine of fish from integrated fish farming [7] and from shellfish [8]. In addition, processed fish products were often found to contain large numbers of LAB, particularly enterococci. Salting processed fish during production may confer a selective advantage on enterococci because these bacteria are among the most tolerance to high salts concentrations [9].
The genus of enterococcus plays an acknowledged role in the development of organoleptic characteristics in fermented foods such as cheeses and sausages [10]. In addition, some enterococci have important implications in the food industry. These bacteria share a number of useful biotechnological traits (e.g. bacteriocins production), which led to earlier applications in fermented foods [2,11]. Bacteriocins are bacterial peptides or proteins that inhibit strains and species that are usually, but not always, closely related to the producing bacteria [12]. Some of these bacteriocins exhibit broad inhibitory spectra and have potential as food preservatives [13].
Some enterococcal strains have been used successfully as probiotics [14,15]. Their success as probiotics has been attributed to factors such as acid and bile resistance, production of antimicrobials and their ability to survive and compete in the gastrointestinal tract. However, enterococci may also be negatively associated with foods due to its possible implication as an indicator of faecal contamination [16]. The question in the safety and acceptability of enterococci in the food and probiotic situation is still heavily debated [10]. It should however be emphasized that some well-defined strains with a long history of safe use are known. Moreover, the particular role and benefits of enterococci in certain food fermentations deserve increased attention in research [17-19].
The objective of this study is to characterize E. faecium strains isolated from Mediterranean fish and to study their bacteriocinogenic activities.
2. Materials and Methods
2.1. Bacterial Strains and Media
LAB strains were propagated aerobically in de ManRogosa-Sharpe (MRS) broth at 30˚C except for lactococal strains, which were grown in M17 broth supplemented with 0.5% (w/v) lactose. Other bacteria were cultured in brain heart infusion (BHI) broth at 37˚C for 24 H.
2.2. LAB Isolation from Wild Fish
Fishes of Pagellus bogarareo were washed with sterile distillate water to remove the unwanted particles. Then, the animals were dissected to remove the digestive tracts in sterile conditions. The digestive tracts were homogenized in NaCl solution (9 g/100 ml) and serially diluted. Two hundred microliters of these dilutions were plated on nutrient agar plates (M17 and MRS) and incubated at 30˚C during 24 to 48 hours. After incubation, colonies were randomly isolated and tested for bacteriocins production against Listeria (L.) ivanovii BUG 496 and Lactococcus (Lc.) garvieae.
2.3. Bacteriocin Assay and Antimicrobial Spectrum
Isolated colonies were inoculated in M17 or MRS broth and incubated for 24 h. After centrifugation, the antibacterial activities of cell-free culture supernatants were measured by the agar-well diffusion method [20], using various strains as indicator cells. The presence of a distinct inhibition zone around the well was considered as positive antagonistic effect. A spectrum of activity was defined against various strains of Gram positives and Gram negatives bacteria (Table 1). E. faecium MMT21 was used as positive control [21].
2.4. Initial Characterization of the Bacteriocin-Producing Strains
The isolates showing bactericidal activities were purified by successive streaks on MRS or M17 plates at 30˚C. The purified cultures were classified according the Gram staining, cellular morphology, catalase and oxidase production. The strains were also examined according to Schleifer and Kilpper-Bälz [22] and further tested for growth on bile esculin azide agar (BEA) media and antibiotic susceptibility by using ATB ENTEROC 5 strips (BioMérieux). Fermentation patterns were determined with API Rapid 50-CHL system (BioMérieux) according to the manufacturer’s instructions. All assays were performed at least in triplicate and the average values were used for analysis.
2.5. Effects of pH, Temperature and Lytic Enzymes on Antimicrobial Activity
The antimicrobial activities of LAB cell-free supernatant after exposure to high temperature (100˚C for 10 min), to pH 3 - 12, or treatment with proteinase K, trypsin, lipase C and a-amylase (10 mg/ml; pH 7) at 37˚C for 30 min were determined using the agar well diffusion assay against the indicator strain Lc. lactis ssp. cremoris ATCC11603 as described above. All assays were performed at least in triplicate.
2.6. Bacteriocin Production, Partial Purification and Molecular Mass Determination
MRS broth was inoculated with over night cultures (1%, v/v) of enterococci strains. Incubation was at 30˚C, without agitation for 30 h, and bacteriocins activity was determined at appropriate intervals and expressed as arbitrary unity/ml (AU∙ml−1).
Bacteriocins produced by our isolates were partially purified according to the two-steps procedure as described previously [21], i.e. ammonium sulphate precipitation and C-18 solid phase chromatography. Fractions with bacteriocin activity were concentrated by freezedrying and re-suspended in distilled water.
Molecular weight of bacteriocin was estimated by Tricine-SDS-polyacrylamide gel electrophoresis (SDS-PAGE) as described previously [23]. The gels were stained with Coomassie Blue, washed carefully and overlaid with L. ivanovii BUG 496 (106 CFU/mL), embedded in BHI soft agar, to determine the position of the active bacteriocin. A low molecular weight marker (Sigma) was used.
2.7. PCR Amplification, DNA Sequencing and Partial Amplified rDNA Restriction Analysis
DNA was isolated, as described previously [24]. The DNA of each bacterial strain was amplified with the specific 16S primers 27f and 1496R described previously [25] to obtain a DNA fragment of 1400 bp corresponding to part of the gene for 16S genomic DNA. PCR was
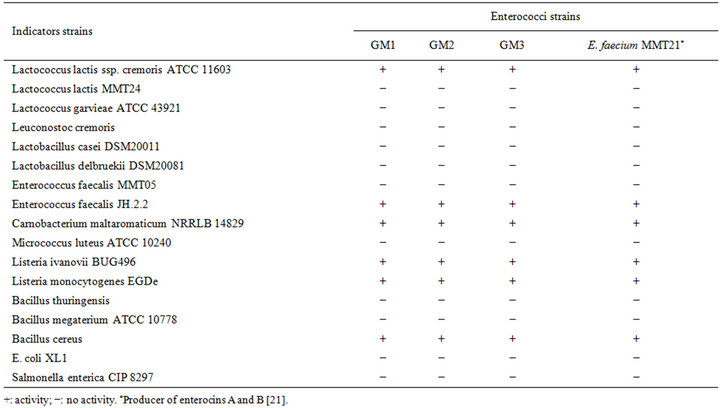
Table 1. Spectrum of activity of cell free supernatant of the three strains.
conducted in a PCR thermal cycler (Techne Flexigene). The conditions used were, initial cycle of denaturation at 95˚C for 5 min, 30 cycles of 92˚C for 45 s, annealing at 58˚C for 1 min, and extension at 72˚C for 1 min, and a final extension cycle of 72˚C for 5 min. PCR products were examined by electrophoresis in 0.8% agarose gel. They were finally purified using QIAquick PCR purification kit (QIAGen) to remove unincorporated primers and nucleotides and subjected to sequencing with an Applied Biosystems 3130 DNA Analyser (Applied Biosystems). The determined nucleotide sequences of 16S rDNA were submitted to GenBank (NCBI, Bethesda, USA) and used for the phylogenetic analyses. The 16S rDNA sequences of other Enterococcus species and E. coli retrieved from GenBank were also used. For phylogenetic analysis, nucleotide substitution rates were determined and a distance matrix tree was constructed by the neighbor-joining method using the CLUSTAL W program.
16S rDNA PCR Amplicons were digested with the restriction endonuclease HaeIII following the manufacturer’s recommendations (Promega). The resulting fragments were analysed by electrophoresis in a 1% agarose gel.
2.8. Identification of the Inhibitory Substances by PCR
PCR amplification was carried out to detect the structural genes of known enterocins (A, B and P) as described previously [21]. The amplified products were separated by electrophoresis in 1% (w/v) agarose gels in 1x TBE buffer. A 100-bp DNA ladder (Invitrogen) was used as a molecular weight marker.
2.9. Nucleotide Sequence Accession Numbers
The 16S rDNA sequences of E. faecium GM1, -2 and -3 were deposited in the NCBI GenBank nucleotide databases under JQ478491, JQ731618 and JQ478493 numbers, respectively.
3. Results
3.1. Isolation of Enterococci Strains from Fish
Three randomly selected strains of enterococci isolated from Pagellus bogarareo exhibited an antilisterial activity (Table 1). All strains are Gram-positive, catalasenegative, oxidase-negative, cocci and were identified according to physiological tests (Table 2). All isolates were thermophilic and grew at 40˚C and 45˚C, but not at 10˚C. They were able to grow at pH 9.6, at 4.5% and 6.5% of NaCl and in BEA medium. They fermented glucose, galactose, mannose, mannitol, lactose and N-acetyl-D-glucosamine. The fermentation of maltose, gentibiose and trehalose was strain-dependent. These results allowed a preliminary identification of our isolates as Enterococcus faecium.
None of the isolates tested demonstrated resistance to ampicillin, penicillin, tetracycline, vancomycin and gentamicin (Table 2). All isolates were resistant to rifam

Table 2. Physiological and biochemical characteristics of isolates.
picin (Table 2). Resistance to rifampicin seems to be widely spread among enterococci [26].
3.2. Identification by 16S Sequencing and Partial Amplified rDNA Restriction Analysis
The bacteriocinogenic LAB isolates were analysed by DNA sequencing to identify the species. The sequence similarity of 16S rDNA between LAB isolates and the other species of enterococci was less than 90%, with the highest degree of relation for E. faecium. 16S rRNAdendrogram was shown in Figure 1. In this tree, enterococci isolates were grouped into two branches. The first branch contained GM1 and GM3 strains, while the second contained GM2 strains. This observation was supported by high bootstrap values. In 16S rRNA nucleotide sequence analysis, GM1 and GM2 strains were found to have 100% similarity to each other and 83% similarity, to GM2 strain.
Enzymatic digestion of the 16S rDNA PCR amplicons with a specific restriction enzyme HaeIII was performed in order to investigate the biodiversity within the enterococci strains from fish viscera. Two different digestion profiles were obtained (Figure 2). E. faecium GM1 and GM3 present the same profile, while GM2 strain gave a
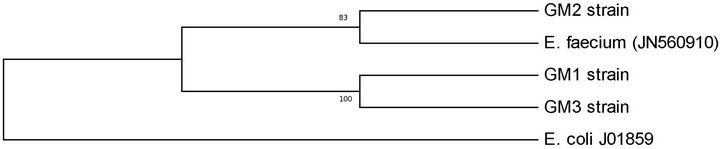
Figure 1. Phylogenetic tree of Enterococcus faecium strains obtained from digestive tract of Tunisian fish, based on 16S rRNA sequences analysis by UPGMA method using MEGA 5.05 software.
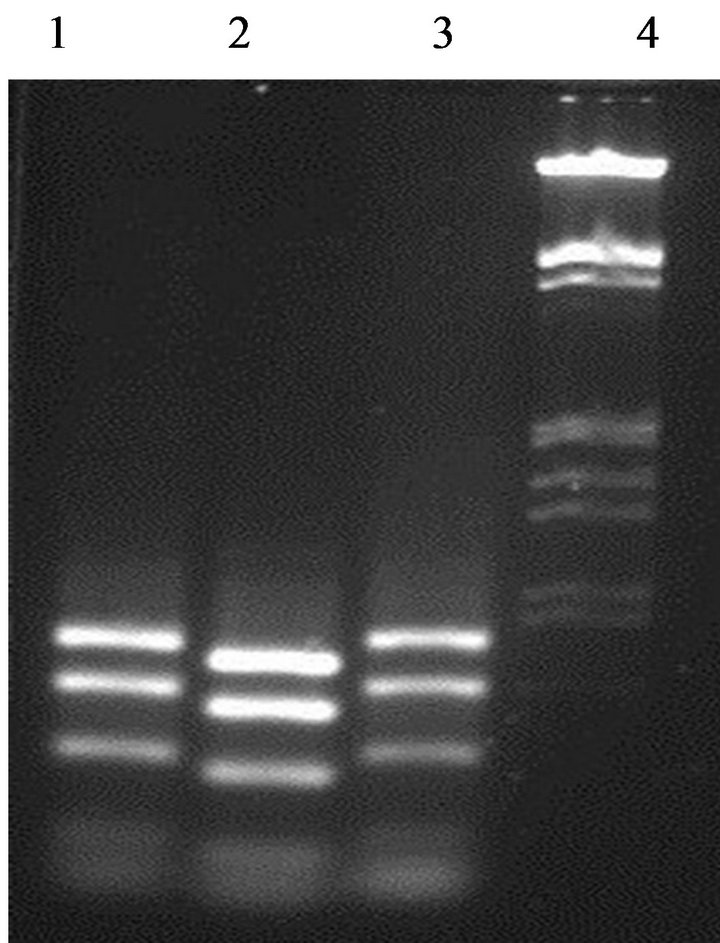
Figure 2. Differentiation between enterococci strains by partial amplicons digested with restriction enzyme HaeIII. Lane 1: strain GM1; Lane 2: strain GM2; Lane 3: strain GM3; Lane 4: DNA marker (100 bp ladder).
different profile. This data confirming that the strain E. faecium GM2 was different to the other enterococci isolates.
3.3. Effect of Enzymes, pH and Heat Treatment on the Antimicrobial Activity
Antagonistic activity of isolates was inactivated by treatment with proteolytic enzymes. This suggested that the inhibitory compound was proteinaeous in nature. Nonproteolytic enzyme has no effect on the activity of the bacteriocin-like inhibitors produced by enterococci isolates. The antimicrobial activity of the free-cell culture supernatant was stables to heat, suggesting that the inhibition observed was due to a low molecular weight protein. In addition, the antimicrobial compound produced by the enterococci isolates was stable at different range of pH, but higher activity was observed at low pH.
3.4. Spectrum of Activity, Bacteriocin Production and Molecular Mass Determination
The cell-free supernatant of the entercocci isolates was tested against a range of Gram-positive and Gram-negative bacteria using the well diffusion assay. Enterococci isolates inhibited the two Listeria strains tested as well as Bacillus cereus (Table 1). A range of LAB was tested, but only C. maltaromaticum, Lc. lactis ssp. cremoris ATCC11603 and E. faecalis JH.2.2 were strongly inhibited by the cell-free supernatant. The bacteriocin likeinhibitors from the isolates displayed a narrow spectrum of antimicrobial activity similar to the enterocins A and B producer, E. faecium MMT21. However, a slight difference in the sizes of the inhibition zones for some indicator strains between enterococci isolates and E. faecium MMT21 was observed (data not shown). Strains of Gram-negative bacteria like E. coli, Salmonella sp. and Pseudomonas aeruginosa were not inhibited. In addition, none of our strains exhibited any antimicrobial activity against Lc. garvieae. The highly sensitive strain for the inhibitory effect of bacteriocins produced by enterococci isolates was L. ivanovii BUG496.
Bacteriocin production occurred in the beginning of the logarithmic growth and increased throughout (Figure 3). The highest level of bacteriocin activity (6100 AU∙ml−1) was recorded after 26 h of growth in MRS broth and remained stable over the next 6 h.
Partial purification of bacteriocins produced by our strains was achieved by ammonium sulphate precipitation and C-18 solid phase chromatography. The fraction showed antimicrobial activity was subjected to TricineSDS-PAGE analysis. A single band migrated to a position corresponding to a peptide of 4500 Da was observed by Coomassie Brilliant Blue staining. This band coincided with a clear inhibition zone of the growth of L. ivanovii BUG 496 (Figure 4).
3.5. Genetic Determination by PCR of the Presence of Enterocins Structural Gene in Enterococcus Isolates
To investigate bacteriocin production, all strains were probed for the presence of the structural genes for enterocins A, B and P. All of the enterococci strains gave a
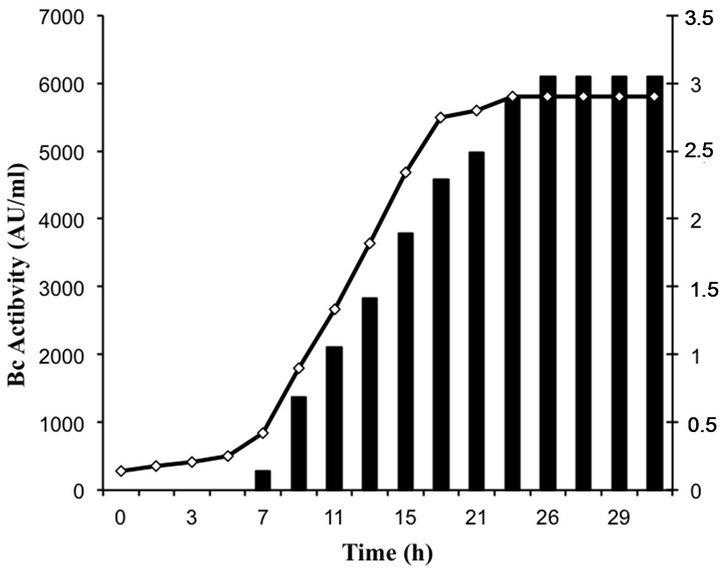
Figure 3. Growth and bacteriocin production of enterococci isolates.
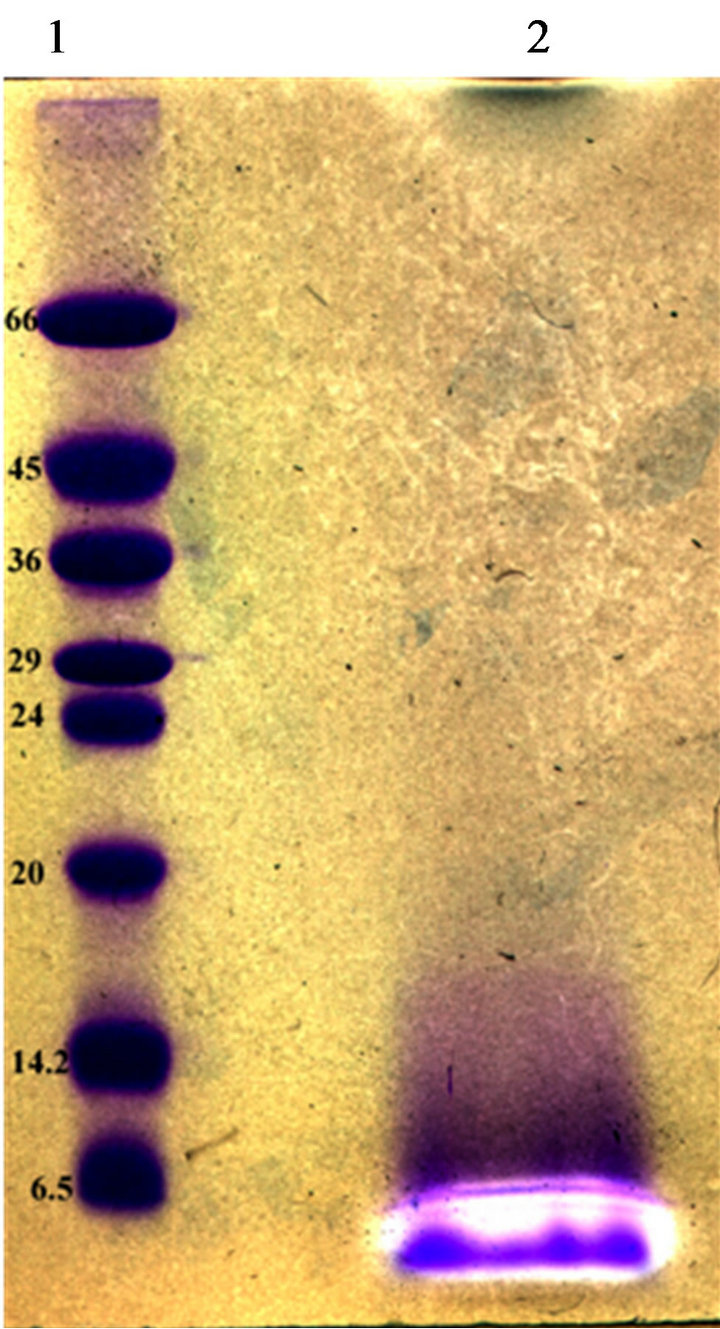
Figure 4. Tricine-SDS–PAGE of bacteriocins from E. Faecium GM2. Inhibition zones indicated the position of the active peptide bands. Lane 1, molecular size standard (size indicated on the left, kDa). Lane 2, partial purified bacterioccins GM02 sample.
positive result for enterocin A (Figure 5). There was no reaction with the enterocin B and P primers. Bacteriocins produced by our strains are thus similar to enterocin A.
4. Discussion
In the present study, three bacteriocinogenic strains were obtained from Pagellus bogarareo viscera. All the isolates were considered as LAB based on their positive Gram reaction, absence of catalase, oxidase and their coccal shape. The primordial microbiological tests show that strains with antibacterial activity have the criteria of E. faecium; since the isolates have been shown to be Gram-positive, catalase negative, to grow at 30˚C and 45˚C and in 6.5% NaCl and to have characteristic carbohydrates fermentation patterns.
Analyses of 16 S rDNA sequences also identified our strains as E. faecium. Considering that E. faecium isolates were obtained from the 10−6 dilution, it seems that it were a dominant species in Pagellus bogarareo intestinal tract and these may suggest their beneficial roles in gut fish. To our knowledge, this is the first characterized E. faecium strains isolated from Tunisian fishes. Other researchers have previously reported E. faecium and E. faecalis to be a member of the bacterial flora in the fish viscera [27], but the effect of their presence is still unknown. Nor is it known if the E. faecium population is the result of an environmental contamination or inherent for the indigenous microflora characterized by a late development. Enterococcus species are highly resistant to adverse conditions and grow well in salt water [28]. Fur-
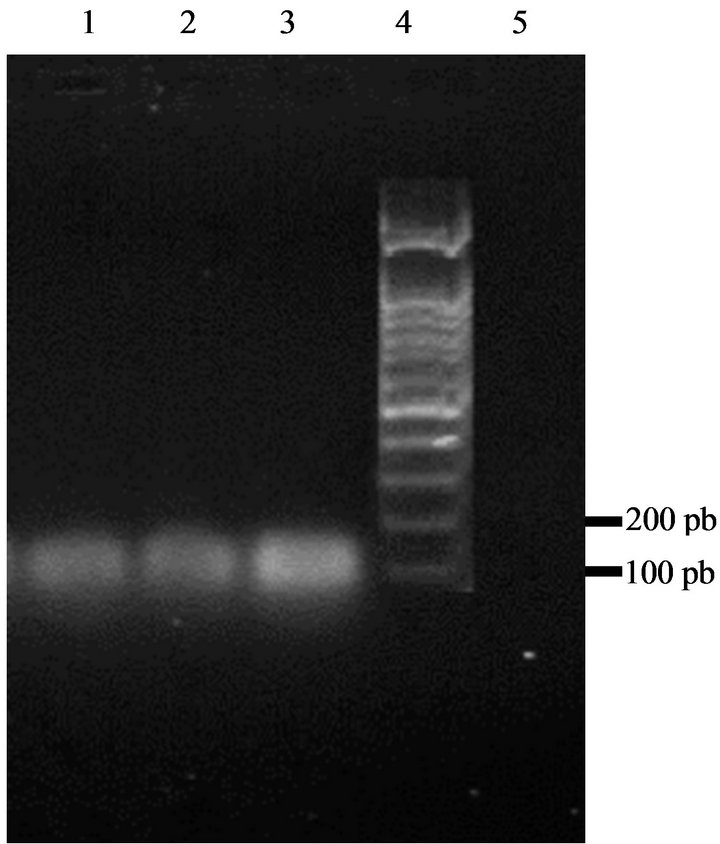
Figure 5. Amplification of DNA of enterococci isolates with specific enterocins primers yielded a 139 bp fragment characteristic for enterocin A. Lane 1: strain GM1; Lane 2: strain GM2; Lane 3: strain GM3; Lane 4: DNA marker (100 bp ladder); Lane 5: control sample (no DNA).
thermore, the indigenous microflora of fish can be founded in fish products and highly influence the quality of the product during processing and storage. Although, more research were needed to consider enterococci as desirable microflora.
The enterococci bacteriocin-producing isolates tolerate bile and did not display vancomycin resistance, an important safety conditions when considering Enterococcus as potential probiotic or food starter culture [16]. It should be noted that bacteriocins production by E. faecium GM01, -02 and -03 were detected even at 15˚C (data not shown). These results were interesting in case the strains are used as protective cultures in chilled foods.
The isolates showed an inhibitory activity against various strains, especially against C. maltaromaticum and L. monocytogenes. C. maltaromaticum was pathogenic for several fish species, including Australian salmonids, carp, rainbow trout, striped bass and channel catfish, and salmon [29]. In addition, it has been reported that some strains of C. maltaromaticum were resistant to several antibiotics widely used in aquaculture [30]. Thus, E. faecium GM1, -2 and -3 could be suggested as probiotic cultures for use in aquaculture. However, safety aspects should be confirmed specially absence of virulence determinants [31].
As expected, none of the Gram-negative indicators strains were inhibited. The isolates produced bacteriocins that were stable at different range of pH with a higher activity at acidic pH values. These results corroborate previous reports referring to LAB strains of other origins that have described a stronger effect of bacteriocins at low pH values [32]. These antagonistic substances did not lose activity after heat treatment and thus similar to most other bacteriocins. Treatment with α-amylase or phospholipase C did not change the antimicrobial activity, suggesting that bacteriocins were not glycosylated or having a lipid moiety.
Three bacteriocin-producing strains were isolated in this study. All three bacteriocins were shown to be stable peptides that conserved activity at all conditions tested. The molecular sizes of the bacteriocin produced by GM02 strain was determined by overlaid tricine-SDSPAGE gel (Figure 4). One antimicrobial peptide bands was recorded with an apparent molecular size of about 4.5 kDa. Thus, enterococci GM02 produce small and hydrophobic bacteriocins within the range of most bacteriocins reported for the genus Enterococcus. Bacteriocin production started at early exponential phase, with maximum production (6100 AU∙ml−1) towards the end of the stationary growth, i.e. after 26 h. Similar result was recorded for bacteriocins produced by Lactobacillus plantarum [33].
In the present study the three enterococci strains GM1, -2 and -3 amplified the expected fragment containing the structural gene of enterocin A. In addition, they were resistant to the culture supernatant of the enterocins A and B producing strains E. faecium MMT21 (data not shown). Based on these results, it’s appeared that strains from fish viscera produce the same bacteriocins to those produced by enterococci isolated from food [34]. Our results confirm that enterocin A was the most commonly detected bacteriocin among E. faecium strains. In addition, our enterocin A producing strains presented a strong anti-Listeria activity, whereas the enterocin-producing food isolate E. faecium MMT21 showed a weaker inhibitory activity. The variability in enterocin A production levels may explained this differences. The structural gene of enterocins B or P was not detected by PCR in the DNA from our isolates. However, it should be mentioned that the presence of multiple enterocin genes is common for a single enterococci strain [22,35].
Partial ARDRA results revealed difference between enterococci isolates from fish viscera. This result suggests genotypic variability in E. faecium from Pagellus bogarareo intestinal tracts. Partial amplified rDNA restriction analysis was a simple and rapid method in species differentiation, However, the conservative nature of 16S rRNA genes limits the discriminatory power of techniques such as ARDRA compared to methods, which utilize differences in the whole genome, such as PFGE and RAPD.
5. Conclusion
In conclusion, we have isolated different E. faecium strains producing enterocin A from fish viscera. These new E. faecium strains could be useful for improving safety of seafood products or probiotics application in aquaculture.
6. Acknowledgements
Sarra Migaw was partially supported by a grant from the Tunisian Ministry of High Education. The second author is grateful to the French Embassy in Tunis for research fellowship.
REFERENCES
- T. J. Eaton and M. J. Gasson, “Molecular Screening of Enterococcus Virulence Determinants and Potential for Genetic Exchange between Food and Medical Isolates,” Applied and Environmental Microbiology, Vol. 67, No. 4, 2001, pp. 1628-1635. doi:10.1128/AEM.67.4.1628-1635.2001
- G. Giraffa, “Enterococci from Foods,” FEMS Microbiology Reviews, Vol. 26, No. 2, 2002, pp. 163-171. doi:10.1111/j.1574-6976.2002.tb00608.x
- W. Chingwaru, S. F. Mpuchane and B. A. Gashe, “Enterococcus faecalis and Enterococcus faecium Isolates from Milk, Beef and Chicken and Their Antibiotic Resistance,” Journal of Food Protection, Vol. 66, No. 6, 2003, pp. 931-936.
- A. S. Valenzuela, N. Ben Omar, H. Abriouel, M. M. Cañamero and A. Gálvez, “Isolation and Identification of Enterococcus faecium from Seafoods, Antimicrobial Resistance and Production of Bacteriocin-Like Substances,” Food Microbiology, Vol. 27, No. 7, 2010, pp. 955-961. doi:10.1016/j.fm.2010.05.033
- M. R. Foulquié-Moreno, P. Sarantinopoulos, E. Tsakalidou and L. De Vuyst, “The Role and Application of Enterococci in Food and Health,” International Journal of Food Microbiology, Vol. 106, No. 1, 2006, pp. 1-24. doi:10.1016/j.ijfoodmicro.2005.06.026
- T. Hagi, D. Tanaka, Y. Iwamura and T. Hoshino, “Diversity and Seasonal Changes in Lactic Acid Bacteria in the Intestinal Tract of Cultured Freshwater Fish,” Aquaculture, Vol. 234, No. 1-4, 2004, pp. 335-346. doi:10.1016/j.aquaculture.2004.01.018
- A. Petersen and A. Dalsgaard, “Species Composition and Antimicrobial Resistance Genes of Enterococcus spp., Isolated from Integrated and Traditional Fish Farms in Thailand,” Applied and Environmental Microbiology, Vol. 5, No. 5, 2003, pp. 395-402. doi:10.1046/j.1462-2920.2003.00430.x
- I. G. Wilson and G. G. McAfee, “Vancomycin-Resistant Enterococci in Shellfish Unchlorinated Waters, and Chicken,” International Journal of Food Microbiology, Vol. 79, No. 3, 2002, pp. 143-151. doi:10.1016/S0168-1605(02)00063-6
- V. J. Harwood, J. Whitlock and V. Withington, “Classification of Antibiotic Resistance Patterns of Indicator Bacteria Bydiscriminant Analysis, Use in Predicting the Source of Fecal Contamination in Subtropical Waters,” Applied and Environmental Microbiology, Vol. 66, No. 9, 2000, pp. 3698-3704. doi:10.1128/AEM.66.9.3698-3704.2000
- C. M. A. P. Franz, M. E. Stiles, K. H. Schleiferc and W.H. Holzapfel, “Enterococci in Foods a Conundrum for Food Safety,” International Journal of Food Microbiology, Vol. 88, No. 2-3, 2003, pp. 105-112. doi:10.1016/S0168-1605(03)00174-0
- F. Leroi, M. R. Foulquié-Moreno and L. De Vuyst, “Enterococcus faecium RZS C5, an Interesting Bacteriocin Producer to Be Used as a Coculture in Food Fermentation,” International Journal of Food Microbiology, Vol. 88, No. 2-3, 2003, pp. 235-240. doi:10.1016/S0168-1605(03)00185-5
- T. R. Klaenhammer, “Genetics of Bacteriocins Produced by Lactic Acid Bacteria,” FEMS Microbiology Reviews, Vol. 12, No. 1-3, 1993, pp. 39-85.
- H. Chen and D. G. Hoover, “Bacteriocins and Their Food Applications,” Comprehensive Reviews in Food Science and Food Safety, Vol. 2, No. 3, 2003, pp. 82-100. doi:10.1111/j.1541-4337.2003.tb00016.x
- L. L. Agerholm, M. L. Bell, G. K. Grunwald and A. Astrup, “The Effect of a Probiotic Milk Product on Plasma Cholesterol, a Meta-Analysis of Short-Term Intervention Studies,” European Journal of Clinical Nutrition, Vol. 54, No. 11, 2000, pp. 856-860. doi:10.1038/sj.ejcn.1601104
- A. Weiss, K. J. Domig and W. Kneifel, “Comparison of Selective Media for the Enumeration of Probiotic Enterococci from Animal Feed,” Food Technology and Biotechnology, Vol. 43, No. 2, 2005, pp. 147-155.
- C. M. A. P. Franz, W. H. Holzapfel, and M. E. Stiles, Enterococci at the Crossroads of Food Safety. International Journal of Food Microbiology, vol. 47, 1999, pp. 1–24. doi:10.1016/S0168-1605(99)00007-0
- M. T. Aymerich, M. Garriga, J. Ylla, J. Vallier, J. M. Monfort and M. Hugas, “Application of Enterocins as Biopreservatives against Listeria innocua in Meat Products,” Journal of Food Protection, Vol. 63, No. 6, 2000, pp. 721-726.
- W. H. Holzapfel, “Appropriate Starter Culture Technologies for Small-Scale Fermentation in Developing Countries,” International Journal of Food Microbiology, Vol. 75, No. 3, 2002, pp. 197-210. doi:10.1016/S0168-1605(01)00707-3
- N. Ben Omar, A. Castro, R. Lucas, H. Abriouel, N. M. K. Yousif, C. M. A. P. Franz, W. H. Holzapfel, R. Pérez-Pulido, M. Martínez-Cañamero and A. Gálvez, “Functional and Safety Aspects of Enterococci Isolated from Different Spanish Foods,” Systematic and Applied Microbiology, Vol. 27, No. 1, 2004, pp. 118-113. doi:10.1078/0723-2020-00248
- T. Ghrairi, M. Manai, J. M. Berjaud and J. Frère, “Occurrence of Anti-Listeria Activity in Lactic Acid Bacteria Strains Isolated from Rigouta, a Traditional Tunisian Cheese,” Journal of Applied Microbiology, Vol. 97, No. 3, 2004, pp. 621-628. doi:10.1111/j.1365-2672.2004.02347.x
- T. Ghrairi, J. Frère, J. M. Berjaud and M. Manai, “Purification and Characterisation of Bacteriocins Produced by Enterococcus faecium from Tunisian Rigouta Cheese,” Food Control, Vol. 19, No. 2, 2008, pp. 162-169. doi:10.1016/j.foodcont.2007.03.003
- K. Schleifer, and R. Kilpper-Bälz, “Transfer of Streptococcus faecalis and Streptococcus faecium to the Genus Enterococcus nom. rev. as Enterococcus faecalis comb. nov. and Enterococcus faecalis comb. nov.,” International Journal of Systematic Bacteriology, Vol. 34, No. 1, 1984, pp. 31-34. doi:10.1099/00207713-34-1-31
- Y. Fleury, M. A. Dayem, J. J. Montagne, E. Chaboisseau, J. P. Le Caer, P. Nicolas and A. Delfour, “Covalent Structure, Synthesis, and Structure-Function Studies of Mesentericin Y 105(37), a Defensive Peptide from Gram-Positive Bacteria Leuconostoc Mesenteroides,” Journal of Biological Chemistry, Vol. 271, No. 24, 1996, pp. 14421- 14429. doi:10.1074/jbc.271.24.14421
- D. G. Anderson and L. L. Mc Kay, “Simple and Rapid Method for Isolating Large Plasmid DNA from Lactis Streptococci,” Applied and Environmental Microbiology, Vol. 46, No. 3, 1983, pp. 549-552.
- W. G. Weisburg, S. M. Barns, D. A. Pelletier and D. J. Lane, “16S Ribosomal DNA Amplification for Phylogenetic Study,” Journal of Bacteriology, Vol. 173, No. 2, 1991, pp. 697-703.
- J. Andrews, J. Ashby, G. Jevons, T. Marshall, N. Lines and R. Wise, “A Comparison of Antimicrobial Resistance Rates in Gram-Positive Pathogens Isolated in the UK from October 1996 to January 1997 and October 1997 to January 1998,” Journal of Antimicrobial Chemotherapy, Vol. 45, No. 3, 2000, pp. 285-293. doi:10.1093/jac/45.3.285
- E. Ringø and F. J. Gatesoupe, “Lactic Acid Bacteria in Fish, a Review,” Aquaculture, Vol. 160, No. 3-4, 1998, pp. 177-203. doi:10.1016/S0044-8486(97)00299-8
- G. Tiecco, “Microbiologia dei vari Prodotti Alimentary di Origine Animale,” In: C. Edagricole, Ed., Igiene e Tecnologia alimentaire, Bologna, 2001, pp. 102-103.
- J. J. Leisner, B. G. Laursen, H. Prevost, D. Djamel and P. Dalgaard, “Carnobacterium, Positive and Negative Effects in the Environment and in Foods,” FEMS Microbiology Reviews, Vol. 31, No. 5, 2007, pp. 592-613. doi:10.1111/j.1574-6976.2007.00080.x
- A. M. Baya, A. E. Toranzo, B. Lupiani, T. Li, B. S. Roberson and F. M. Hetrick, “Biochemical and Serological Characterization of Carnobacterium spp. Isolated from Farmed and Natural Populations of Striped Bass and Catfish,” Applied and Environmental Microbiology, Vol. 57, No. 11, 1991, pp. 3114-3120.
- I. Kuhn, A. Iversen, M. Finn, C. Greko, L. G. Burman, A. R. Blanch and X. Vilanova, A. Manero, H. Taylor, J. Caplin, L. Domínguez, I. A. Herrero, M. A. Moreno and R. Möllby, “Occurrence and Relatedness of VancomycinResistant Enterococci in Animals, Humans, and the Environment in Different European Regions,” Applied and Environmental Microbiology, Vol. 71, No. 9, 2005, pp. 5383-5539. doi:10.1128/AEM.71.9.5383-5390.2005
- H. Blom, T. Katla, H. Nissen and H. Holo, “Characterization, Production, and Purification of Carnocin H, a Bacteriocin Produced by Carnobacterium 377,” Current Micryobiology, Vol. 43, No. 4, 2001, pp. 227-231. doi:10.1007/s002840010292
- S. D. Todorov and L. M. T. Dicks, “Screening for Bacteriocin-Producing Lactic Acid Bacteria from Boza, a Traditional Cereal Beverage from Bulgaria Comparison of the Bacteriocins,” Process Biochemistry, Vol. 41, No. 1, 2002, pp. 11-19. doi:10.1016/j.procbio.2005.01.026
- C. M. A. P. Franz, M. J. van Belkum, W. H. Holzapfel, H. Abriouel and A. Gálvez, “Diversity of Enterococcal Bacteriocins and Their Grouping in a New Classification Scheme,” FEMS Microbiology Reviews, Vol. 31, No. 3, 2007, pp. 293-310. doi:10.1111/j.1574-6976.2007.00064.x
- C. Herranz, P. Casaus, S. Mukhopadhyay, J. M. Martinez, J. M. Rodriguez, I. F. Nes, E. Herrandez and L. M. Cintas, “Enterococcus faecium P21, a Strain Occurring Naturally in Dry-Fermented Sausages Producing the Class II Bacteriocins Enterocin A and Enterocin B,” Food Microbiology, Vol. 18, No. 2, 2001, pp. 115-131. doi:10.1006/fmic.2000.0382
NOTES
*Corresponding author.

