American Journal of Plant Sciences
Vol.3 No.8(2012), Article ID:22189,4 pages DOI:10.4236/ajps.2012.38135
Fungal Isolate “KMI” Is a New Type of Orchid Mycorrhizal Fungus
![]()
Graduate School of Life Environmental Sciences, Kyoto Prefectural University, Kyoto, Japan.
Email: s810631049@kpu.ac.jp
Received May 5th, 2012; revised June 1st, 2012; accepted June 10th, 2012
Keywords: KMI; Orchid Mycorrhizal Fungus; Paphiopedilum thailandense; Protein Analysis, Ribosomal DNA Sequence
ABSTRACT
We analyzed an isolate of fungus that seems to be a new type orchid mycorrhizal fungus, “KMI (refers to Kyoto-Matsubara-Ishii)”, obtained from the roots of Paphiopedilum thailandense Fowl. KMI has hyphal branching into right angle similar to Rhizoctonia-like fungi, but its spore formation resembled Nectoria, which is known as a teleomorph of Fusarium rather than Rhizoctonia. Its ribosomal DNA sequences of 18S and ITS have no similarity with any known fungal species. Proteins in molecular weight of 53 and 24 kDa, which are common to mycorrhizal fungi, were detected in KMI. When KMI was inoculated onto orchids, however it didn’t form pelotons or coils, but the hyphae in the root tissues were observed and the tissues were not decayed. No significant symptoms of Rhizoctonia or Fusarium disease, however, were developed on tomato and cucumber plants. The evidences suggest that KMI is a new type of orchid mycorrhizal fungus.
1. Introduction
It is known that orchid mycorrhizal fungi contribute to orchid seed germination, and generally colonize the root tissues near from the surface of adult orchid plant roots. Bernard [1] observed Rhizoctonia repens (teleomorph: Tulasnella deliquescens) in roots of Goodyera repens. Rhizoctonia-like orchid mycorrhizal fungi can be classified into Thanatephorus (=Corticium), Ceratobasidium, Ypsilonidium, Sebacina, Tulasnella and other groups based on the morphology of their teleomorph [2]. They can be distinguished from other fungi by their hyphal branching at right angle and the septa forming near the branching point.
Fungal species, which can make orchid mycorrhiza, are not only Rhizoctonia-like species. Kusano [3] discovered that Armillaria mellea can have symbiotic relation with Gastrodia elata. It turned out that not only G. elata but Galeola septentrionalis [4] and Gastrodia cunninghamii [5] are A. mellea’s host. These 3 species of orchids, which have no chlorophyll, are all depending on the fungi for nutrients.
In this study, we aimed to identify an isolate that will be a new type orchid mycorrhizal fungus obtained from Paphiopedilum thailandense roots.
2. Materials and Methods
2.1. Isolation of KMI
Roots of P. thailandense obtained from Kyoto Prefectural Botanical Garden were cut 3 cm from the apex, and were surface-sterilized for 12 minutes in antibiotic solution (700 ppm chloramines-T, 5.6 ppm streptomycin and 2.1 ppm chloramphenicol). Then, they were homogenized by a glass mortar and pestle in a clean bench, placed on potato dextrose agar media (PDA; Eiken Chemical Co., LTD.), and incubated in dark at 25˚C for 3 days.
2.2. Microscopic Examination of KMI
In a clean bench, a small volume of 20% potato dextrose media was dropped on glass slide with two holes. The slide was placed in a 9 cm plastic Petri dish, then hyphae of KMI were added. The slide was covered with a cover glass before observing fungal growth under the optical microscopy (×400) after the same incubation as mentioned above.
2.3. DNA Sequencing
KMI was inoculated on 20 ml of 20% potato dextrose media and incubated in 25˚C for 3 days.
DNA was isolated from the mycelium of KMI dried with liquid nitrogen using the DNAeasy Plant Mini Kit (Qiagen, Tokyo, Japan) according to the manufacturer’s instructions.
The DNA solution was used as the template for PCR. The oligonucleotide primers used for 18S rDNA partial sequence specific PCR were NS1: 5’-GTAGTCATATGCTTGTCTC-3’, NS4: 5’-CTTCCGTCAATTCCTTTAAG-3’, and ITS rDNA specific PCR were ITS4: TCCTCCGCTTATTGATATGC-3’, ITS5: 5’-GGAAGTAAAAGTCGTAACAAGG-3’ [6] made by Proligo (Japan). The reaction mixture consisted of 1 μl of template solution, 2.5 μl of each primer (2 pmol), KOD Dash (2.5 U/μl, TOYOBO) as taq polymerase, 5 μl of dNTPs (2 mM dATP, dGTP, dCTP, dTTP each, TOYOBO) and 5 μl of 10 × reaction buffer (TOYOBO). Sterilized distilled water was added to increase the volume to 50 μl.
The PCR cycling conditions were one cycle of 94˚C for 3 min, 30 cycles of 94˚C for 30 sec, 50˚C for 2 sec, and 74˚C and 30 sec, and final extension step at 20˚C for 3 min.
The PCR products were purified by GENECLEAN Kit (Q-BIOgene) according to the manufacture’s instruction. Purified PCR products were used in sequencing reactions with the same primers using a BigDye Terminator v3.0 Cycle Sequencing Kit (Applied Biosystems). Sequencing was performed on Genetic Analyzer 310 (Applied Biosystems).
Sequences were searched on the nucleotide sequence database using BLAST program (http://www.ebi.ac.uk/ blastall/). Then, phylogenic analysis was conducted by using ClustalW (http://clustalw.ddbj.nig.ac.jp/top-j.html) and TreeView
(http://taxonomy.zoology.gla.ac.uk/rod/treeview.html).
2.4. Extraction and Purification of Proteins
It is known that arbuscular mycorrhizal fungi (AMF) contain proteins where molecular weights are 53 and 24 kDa [7]. To check whether identified orchid mycorrhizal fungi and KMI have those proteins, a modified procedure of Shannon et al. [8] was used to extract and partially purify proteins from KMI, AMF such as Glomus clarum, and 3 identified orchid mycorrhizal fungi such as Screrotium tulipalum (NBRC 6168), Thanatephorus cucumeris (NBRC 101554) and Rhizoctonia candida (NBRC 7033). Each fungus were homogenized with 0.05 M Tris buffer, pH 7.5, containing 0.05 M NaCl by a mortar and pestle. The homogenates were filtered through cheesecloth, and ammonium sulfate was added to 35% saturation.
After standing overnight at 4˚C, the liquid was centrifuged at 8000 g for 30 min. The supernatant liquid was made up to 95% ammonium sulfate saturation. After standing overnight at 4˚C, the liquid was centrifuged. The residues were dissolved in 2 ml of 0.005 M Tris buffer, pH 7.0. After centrifugation, the supernatant liquid was dialyzed against 0.005 M Tris buffer, pH 8.0, containing 0.1 M KCl. The dialyzate was centrifuged. The supernatant liquid was used as protein samples.
2.5. SDS-PAGE of Protein
SDS-PAGE was carried out to the methods of Laemmli [9] in order to check whether known orchid mycorrhizal fungi and KMI contain proteins with molecular weights of 53 and 24 kDa.
Each protein samples from KMI, G. clarum, S. tulipalum, T. cucumeris and R. candida were analysed. After the SDS-PAGE, bands on the gel were analyzed by the iMeasureScan software (iMeasure Inc.).
2.6. Inoculation of KMI in Plants
In order to confirm that KMI is a mycorrhizal fungus, it was inoculated on several kinds of orchids. Because P. thailandense, the host of KMI is one of rare species that can not be propagated easily and both of its seeds and plants were unavailable, mericlone cultured plants of two kinds of Phalaenopsis (code: PATO79 “Memorial Day” and code: PATOB1 “Ice Lady”), which have resemblance of morphologic characteristics and Miltonia (code: M203 ENZAN LADY “Stork Feather”) were used for the experiment (These orchids were obtained from Mukoyama Orchids Co., Ltd.).
Eighty ml of IH base media [10] were poured into 800 ml jars. After the media were solidified, one orchid plant was transplanted in each jar. Half of them were inoculated with 5 ml of KMI cultural solution (cultured by 20% potato dextrose liquid media for 3 days in 25˚C). After incubated at 28˚C for a week, some of their roots were cut and stained using the method of Phillips and Hayman [11] and observed by using a light microscope.
To check whether KMI is pathogenic or not to vegetables which are sensitive to Rhizoctonia and Fusarium, 5 plants each of tomato (Solanum lycopersicum L.) and cucumber (Cucumis sativus L.) were inoculated with KMI. That is, their seeds were sown in sterilized vermiculite. A month after their germination, the plants were individually transplanted into plastic pots with sterilized vermiculite. Each plant received 5 ml of KMI cultural solution, containing the fungus mycelia.
3. Results
On each PDA plate, KMI formed a white colonie. No other fungal colonies were observed. Diameter of KMI hyphae was ranged from 1.5 to 2.5 μm, and branched to right angle (Figure 1(a)). It seldom made septa. Its spores were oval shape in size of 2 μm × 4 μm and had two nuclei each (Figure 1(b)).
KMI had no significant similarity to any reported fungal species in both of 18S rDNA partial sequence:
5’-CTCATTATATAAGTTATCGTTTATTTGATAGTACCTTACTACTTGGATAACCGTGGTAATTCTAGAGCTA ATACATGCTAAAAATCCCGACTTTGGAAGGGATGTATTTATTAGATTAAAAACCAATGCCCTTCGGGGCTCCTTGGTGATTCATGATAACTCCTCGAATCGCATGGCCTTGTGCCGGCGATGGTTCATTCAAATTTCTTCCCTATCAACTTTCGATGTTTGGGTATTGGCCAAACATGGTTGCAACGGGTAACGGAGGGTTAGGGCTCGACCCCGGAGAAGGAGCCTGAGAAACGGCTACCACATCCAAGGAAGGCAGCAGGCGCGCANATTACCCAAATCCTAACACGGGGAAGGTAGTGACAATAAATAAACAATACCGGGCTCATTGAAGTCTGGTAATTGGAATGAGTACAATCTAAATCCCTTAACGAGGATCCATTGGAGGGCAAGTCTGGTGCCAGCAGCCGCGGTAATTCCAGCTCCAATAGCGTATATTAAAGTTGTTTGCAGTTAAAAGCTCGTAGTTGGACCTTGGGNTGGGTCGATCGGTCCGCCTCTGGGTGTGCACCGGTCGGCTCGTCCCTTCT GC-3’ (Figure 2) and ITS rDNA partial sequence:
5’-CCCCTGTGAACTATACCGATTTGTTGCTTCGGCGGTGCCCCTCGCTTTCGCGGCCAGGGCCC GCCAGA GGACCCAAATTCTTTGTTTTTTTGTATTCGTTTTATATTCTTCTGAGTGGAATTTTTTAAATAAAAAATTAAAACTTTCAACAACGGATCTCTTGGCTCTGGCATCGATGAAGAACGCAGCGAAATGCGATAAGTAATGTGAATTGCAGAATTCAGTGAATCAT
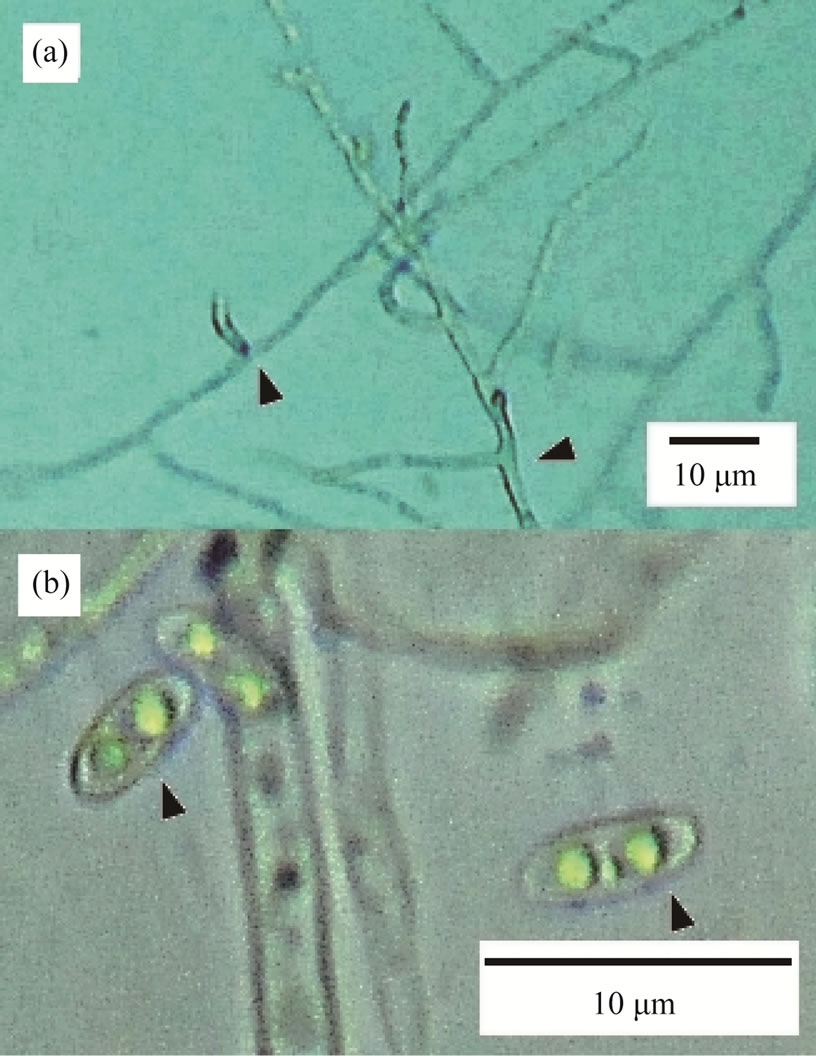
Figure 1. Morphological characteristics of KMI. (a) The arrows show the right-angle branching points; (b) KMI spores have two nuclei.

Figure 2. Phylogenetic analysis of KMI by the 18S rDNA sequence. The white and gray box shows KMI and species which belongs to Rhizoctonia, respectively.
CGAATCTTTGAACGCACATTGCGCCCGCCAGTATTCTGGCGGGCATGCCTGTTCGAGCGTCATTTCAACCCTCAAGCCCCCGGGCTTGGTGTTGGGGATCGGCACAAGGCCCCTCGTGGGCCCGCCGTCCCCCAAATGCAGTGGCGGTCACGTCGCAGCCTTCTATGCGTAGTAGCAACACCTCGCACTGGAGCGCGACGCGGCCACGCCGTAAAACCCCCGACTTTTTTCTGGTTGACCTCGAATCAGGTAGGACTACCCGCTGAACTTAAGCATATC-3’ (Figure 3).
Proteins in molecular weights of 53 and 24 kDa were detected in G. clarum, KMI and all of three orchid mycorrhizal fungi (Figure 4).
The analysis of iMeasureScan software showed the fact that KMI have both molecular weights of proteins.
KMI infected all three types of orchids (Figure 5). Hyphae were observed in tissues near the surface of the roots and they were not decayed. However KMI didn’t form pelotons or coils, not only in potato dextrose media but also when inoculated on orchid roots, the morphological characteristics of hyphae branching to right angle and seldom makes septa, were observed (Figure 5(c)). Although tomato growth was decreased after two weeks from inoculation of KMI, no disease symptoms of Rhizoctonia or Fusarium were observed on both plants of tomato and cucumber (Table 1).
4. Discussion
KMI has hyphal branching into right angle, which looks similar to Rhizoctonia species, and can infect the surface tissues of orchid roots. Even when the hyphae penetrated

Table 1. Effect of the inoculation of KMI on the growth of tomato and cucumber plants.

Figure 3. Phylogenetic analysis of KMI by the ITS rDNA sequence. The white box shows KMI. The Dark grey boxes show species which belong to Rhizoctonia. Fungal sp. ARIZ B394 and B393 on the light grey boxes show species which will help germinate seeds of a tropical tree, Cecropia insignis.
into the root tissues, the root cells were not decayed. The evidences show that KMI is an orchid mycorrhizal fungus. Interestingly, when the similarity of 18S rDNA among KMI and Rhizoctonia species was analyzed, Sclerotium rolfsii (one of the Rhizoctonia species) and KMI existed in a same cluster. But the similarity between them was only 88% and the morphological characteristics of KMI spores were different from those of Rhizoctonia [12]. It seems to resemble Nectoria, which is known as a teleomorph of Fusarium rather than Rhizoctonia. In addition, KMI didn’t form pelotons or coils in the root cells.
Since some of the Fusarium species are known as endophytes in protocorms or roots of orchid plant and Vujanovic et al. [13] reported that an isolate of such Fusarium stimulated seed germination and protocorm formation in Cypripedium reginae, and suggested the

Figure 4. The digitize image of proteins extracted from some kinds of mycorrhizal fungi. (a) KMI; (b) Glomus clarum; (c) Sclerotium tulipalum (NBRC 6168); (d) Thanatephorus cucumeris (NBRC 101554); (e) Rhizoctonia candida (NBRC 7033). White and black arrows show 53 and 24 kDa proteins, respectively.
potential of some Fusarium species to have symbiotic relation with orchid plants. But KMI rDNA shows low similarity with known Fusarium species in both 18S (<94%) and ITS (<92%). It seems that KMI doesn’t belong among the group of Fusarium.
Fungal sp. ARIZ B394, which had highest similarity (96%) of the ITS sequence of KMI, is reported as an endophyte of tropical tree Cecropia insignis [14] and may help germination of their seeds. Thus, KMI can be a fungal species that can have symbiotic relation with orchid plants. In addition, the fact that the inoculation of KMI on tomatoes and cucumbers did not cause the symptom such as wilt suggested that KMI was not a pathogenic fungus.

Figure 5. Orchid roots inoculated by KMI, one week after the inoculation. (a) Phalaenopsis “Memorial Day” roots; (b) Phalaenopsis “Ice Lady” roots; (c) Miltonia “Stork Feather” roots. The arrow shows the right-angle branching point. The “h” in each figure refers to “hypha”.
Proteins in molecular weight of 53 and 24 kDa were common to AMF and orchid mycorrhizal fungi. Because KMI also has proteins in same molecular weights, those can be used for symbiosis. It will be interesting to search the mechanism of symbiosis between orchid plants and KMI.
In this study, we observed formation of mycorrhiza on Phalaenopsis and Miltonia roots, because P. thailandense was so rare that the plants without any infections on their roots were unavailable. The fact that KMI infected these different types of orchids shows that not only P. thailandense but also some other kinds of orchids can be host of KMI.
Since the seeds of P. thailandense were unavailable, it is necessary to check how KMI effects at the germination stage of P. thailandense seeds.
5. Acknowledgements
We are grateful to Dr. Chantal Hamel of Saskatchewan University for her critical reviews on the manuscript. We wish to thank Dr. Andre Freire Cruz for his advice about DNA analysis. We wish to thank Mr. Sukeyasu Namura of Kyoto Prefectural Botanical Garden, Kyoto, Japan and Mr. Takehiko Mukoyama of Mukoyama Orchids Co., LTD., Japan for providing us with orchid plants.
REFERENCES
- N. Bernard, “Recherches Experimentales sur les Orchidees,” Revue Générale de Botanique, Vol. 16, 1904, pp. 405-451, 458-476.
- R. S. Currah and C. Zelmer, “A Key and Notes for the Genera of Fungi Mycorrhizal with Orchids and a New Species in the Genus Epulorhiza,” Reports of Tottori Mycological Institute, Vol. 30, 1992, pp. 43-59.
- S. Kusano, “Gastrodia Elata and Its Symbiotic Association with Armillaria Mellea,” Journal of the College of Agriculture, Vol. 4, 1911, pp. 1-66.
- M. Hamada, “Studien über die Mykorrhiza von Galeola Septentrionalis Reichb. F—Ein neuer Fall der Mykorrhiza-Bildung Durch Intraradicale Rhizomorpha,” Japanese Journal of Botany, Vol. 10, 1939, pp. 151-211.
- E. O. Campbell, “The Mycorrhiza of Gastrodia Cunninghamii Hook. F,” Transactions of the Royal Society New Zealand, Botany, Vol. 1, 1962, pp. 289-296.
- T. J. White, T. Bruns, S. Lee and J. W. Taylor, “Amplification and Direct Sequencing of Fungal Ribosomal RNA genes for Phylogenetics,” In: M. A. Innis, D. H. Gelfand, J. J. Sninsky and T. J. White, Eds., PCR Protocols: A Guide to Methods and Applications, Academic Press, Inc., New York, 1990, pp. 315-322.
- T. Ishii, T. Ikeda, K. L. Rutto, A. F. Cruz, I. Matsumoto and K. Kadoya, “Proteins Related to the Mechanism of Symbiosis between Vesicular-Arbuscular Mycorrhizal Fungi and Plants,” Journal of the Japanese Society for Horticultural Science, Vol. 68, No. 2, 1999, p. 229.
- L. M. Shannon, E. Kay and J. Y. Lew, “Peroxidase Isozymes from Hhorseradish Roots,” Journal of Biological Chemistry, Vol. 241, No. 9, 1966, pp. 2166-2172.
- U. K. Laemmli, “Cleavage of Structural Proteins during Assembly of the Head of Bacteriophage T4,” Nature, Vol. 227, No. 5259, 1970, pp. 680-685. Hdoi:10.1038/227680a0
- T. Ishii and S. Horii, “Establishment of Axenic Cultural Techniques of Arbuscular Mycorrhizal Fungi,” Horticultural Research, Vol. 6, No. 2, 2007, p. 136.
- J. M. Phillips and D. S. Hayman, “Improved Procedures for Clearing and Staining Parasitic and Vesicular-Arbuscular Mycorrhizal Fungi for Rapid Assessment of Infection,” Transactions of the British Mycological Society, Vol. 55, No. 1, 1970, pp. 158-161. Hdoi:10.1016/S0007-1536(70)80110-3
- Y. Uetake, A. Ogoshi and S. Hayakawa, “Observation of Teleomorphs of Rhizoctonias (Thanatephorus orchidicola and Tulasnella deliquescens) Isolated from Orchids,” Memoirs of the Faculty of Agriculture, Vol. 22, No. 2, 1999, pp. 121-125.
- V. Vujanovic, M. St-Arnaud, D. Barabé and G. Thibeault, “Viability Testing of Orchid Seed and the Promotion of Colouration and Germination,” Annals of Botany, Vol. 86, No. 1, 2000, pp. 79-86. Hdoi:10.1006/anbo.2000.1162
- J. M. U’Ren, J. W. Dalling, R. E. Gallery, D. R. Maddison, E. C. Davis, C. M. Gibson and A. E. Arnold, “Diversity and Evolutionary Origins of Fungi Associated with Seeds of a Neotropical Pioneer Tree: A Case Study for Analysing Fungal Environmental Samples,” Mycological Research, Vol. 113, No. 4, 2009, pp. 432-449.

