Open Journal of Immunology
Vol.08 No.04(2018), Article ID:89346,10 pages
10.4236/oji.2018.84006
In Silico Analysis of Cross Reactivity between Lipocalin of Domestic Animals*
Múnera Marlon1, Sanchez Andres1, Sanchez Jorge2, Emiliani Yuliana1
1Medical Research Group (GINUMED), Universitary Corporation Rafael Nuñez, Cartagena, Colombia
2Group of Clinical and Experimental Allergy (GACE), IPS Universitaria, University of Antioquia, Medellín, Colombia
Copyright © 2018 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY 4.0).
http://creativecommons.org/licenses/by/4.0/



Received: October 26, 2018; Accepted: December 21, 2018; Published: December 24, 2018
ABSTRACT
Lipocalins are one of the groups of allergens derived from domestic animals with clinical importance for the development of allergic responses. They have been characterized in different animals. With allergenic capacity characterized, little is known about the epitopes involved in allergic responses. Here, potential antigenic regions involved in cross-reactivity among lipocalins were explored through bioinformatics tools. The amino acid sequences of several lipocalins from different domestic animals (mouse, dog, cat, bull, hamster, horse and pig) were used to determine the degree of kinship by phylogenetic studies. Groups with highest phylogenetic relation were obtained by using MEGA software. 3D models of lipocalins not reported in the protein data bank were modeled by homology to identify potential antigenic regions compromised in the cross-reactivity of this group of allergens. The alignment of the entire database of allergenic lipocalins and the inferred maximum likelihood tree segregate lipocalins into five monophyletic clades (referenced here as A, B, C, D and E). According to the multiple pairing analyzes, group C (Fel d 4, Rat n 1 and Equ c 1) showed the highest degree of identity among their amino acid sequences (58%). The analysis of conserved and exposed residues showed that group C shares three antigenic regions that could potentially contribute to its cross-reactivity. Potential antigenic sites were identified for the generation of cross-reactivity between the different lipocalins analyzed in this study. These studies support the need to carry out directed mutagenesis tests to confirm their relevance in the allergenic capacity of lipocalins.
Keywords:
Bioinformatics, Immunology, Prediction, Epitope, Allergen, Antigen

1. Introduction
The WAO defines the allergy as “A hypersensitivity reaction initiated by immunological mechanisms” [1] . This can be mediated by antibodies or cells, where, in most cases, the antibody responsible is the immunoglobulin E (IgE). The antigens that trigger allergies are defined as allergens, and they are structurally variable molecules, usually linked to a carbohydrate, with IgE binding capacity [2] [3] . Different genetic factors predispose to these diseases and patients may be sensitized to different sources of allergens. Allergic diseases associated with environmental allergens include allergic asthma, rhinitis, conjunctivitis and atopic dermatitis.
An example of these allergens is lipocalins, which represent the most important group of allergens coming from furry animals. In addition, the growing number and diversity of pets in homes has allowed the increase in lipocalin exposure, leading to an increase in sensitization [4] [5] . The lipocalins are a numerous groups of proteins present in vertebrate and invertebrate animals, plants and bacteria, with a great variety of structures and functions between the different species. These biomolecules are relatively small, with an approximate size between 150 - 250 amino acid residues in their primary structure which have the capacity to fulfill different functions, among them; transporter of small hydrophobic molecules such as retinol and binding to surface receptors [4] [5] . The members of this family have been characterized according to their sequence or structure, including a large number of proteins. Within the lipocalins a low degree of conservation is seen in the primary sequences, in some comparisons even values lower than 20% identity [5] [6] [7] [8] .
The identification of allergenic lipocalins from domestic animals has been increasing in the last decade and among patients allergic to pets, co-sensitization to different animals is frequent [9] . Although lipocalins as panallergens could explain the co-sensitization to different pets, little has been studied about the different epitopes involved in cross-reactivity due to these proteins. In this work, using bio-computational tools, we identify different antigenic regions that may be involved in the cross-reactivity between lipocalins.
2. Materials and Methods
2.1. Selection of Lipocalins and Alignment
The amino acid sequences of lipocalins from seventeen domestic animals were selected based on allergenic capacity reported. The sequences were obtained from the uniprot database (https://www.uniprot.org/) (Table 1). Sequences that were reported in the WHO/IUIS Allergen Nomenclature Sub-Committee (http://www.allergen.org) and that presented a complete sequence were used. All the lipocalins that fulfilled this criterion were chosen, independent of the animal source where it came from. Identity grade between lipocalins used in this study was determined by using praline web server (http://www.ibi.vu.nl/) [10] . Parameters to perform alignment were set up to use BLOSUM62 as exchange matrix. Iterations used were 3 with an E-value of 0.01.
2.2. Phylogenetic Analysis
The program Molecular Evolutionary Genetic Analysis (MEGA) version 7 was used for the construction of the trees, using the method of reconstruction of Neighbor-Joining with support by Bootstrap with 500 replications as a measure of reliability and robustness under the assumption of minimal evolution in the topology, this model uses a comparative matrix to find the similarity between amino acids of seventeen sequences to establish the evolutionary proximity between the species. The matrix was constructed with all amino acid sequences of lipocalins retrieved from uniprot database and reported in WHO/IUIS Allergen Nomenclature Sub-Committee (http://www.allergen.org). Thus, the more positive identity values are found among the sequences, the greater their relationship will be and they will be located in closer positions in the tree. All empty spaces were eliminated (full deletions). From the global comparison and the homologies, the sum of the length of branches (SBL) will be presented, which will determine the number of nodes and the position of the same, including the “clusters” of the evolutionarily closest sequences. Due to the number of sequences used, no phylogenetic sub-analyzes were performed. The alignment for the phylogenetic analysis was carried out through the CLUSTAL W. program, which performs alignments.
Table 1. Allergens and allergenic sources.
2.3. Construction of 3D Models
The models of those lipocalins not reported in the protein data bank, were made by homology. The Swiss-model server (https://swissmodel.expasy.org/) was used. The quality of the models was analyzed by ProSA-web. The models were refined in Deep-View (energy minimization and rotamer replacements). Its quality was evaluated by several tools, including the Ramachandran graphs, WHATIF, QMEAN4 index and energy values (GROMOS96 force field). The relative values of the area of accessible solvent (r-ASA) were determined by ASA-view. The lipocalin sequences were aligned to identify conserved residues. Those preserved and the residues accessible to the solvent (rASA > 0.25) were located in the 3D model to identify pooled areas (>4 residues) and possible cross-reactivity.
3. Results
3.1. Phylogenetic Analysis
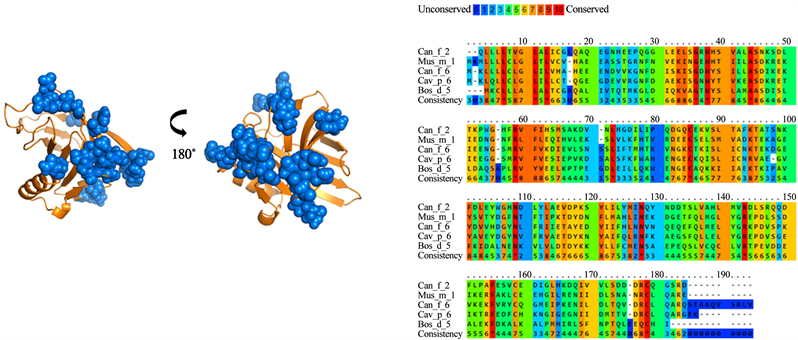
A total of 17 sequence were included in the analysis with 152 positions in the final dataset. The sum of the branch length for an optimal tree was 16.5. When analyzed the lipocalin sequences, it was found that they formed five nodes with the highest phylogenetic relationship among them. According to the analyzes, group A contains the highest number of phylogenetically related lipocalins including Cav p 6, Can f 6, Bos d 5, Mus m 1 and Can f 2. Meanwhile, the group D, threw only two members, Bos d 2 and Can f 4. The group A presents the greatest relationship among the groups (Figure 1).
Figure 1. Phylogenetic tree based on the amino acid sequences of the lipocalins studied. The formation of five clades (A-E) with the highest degree of identity is observed (96% for clade C). The sum of the branch length for an optimal tree was 16.5. The evolutionary distance was computed using the Poisson correction method.
3.2. Construction of 3D Models
The 3D models of those lipocalins not reported in protein data bank were constructed by modeling homology (with the exception of Bos d 2, Can f 2 and Equ c 1, were retrieved from SDAP database (http://fermi.utmb.edu/ ). All generated models show classic folding of lipocalins, following the pattern of eight antiparallel β strands and an α helix, which help to form a cavity for the union of lipid ligands (Figure 2). The models were used to identify residues exposed on the surface and conserved in the different lipocalins of the phylogenetic groups formed.
3.3. Identification of Potential Cross-Reactive Antigenic Sites
Multiple alignments of the lipocalins belonging to the different groups obtained from the phylogenetic analyzes were made. Lipocalins from Group A lipocalins have 30% identity between their amino acid sequences (Figure 3). A total of 20 residues were identified and conserved among the analyzed lipocalins (Table 2), which form two antigenic patches common among the group A.
Figure 2. 3D models of lipocalins from different biological sources used in the study. (a)-(d) Lipocalins are organized according to clades in phylogenetic tree. A structural conservation characteristic of this protein family is observed. *Indicated lipocalins with 3D resolved experimentally.
Figure 3. Analysis of group A lipocalins. Blue balls indicate antigenic residues predicted conserved and exposed on surfaces among lipocalins.
Table 2. Residues conserved among lipocalin groups with antigenic potential.
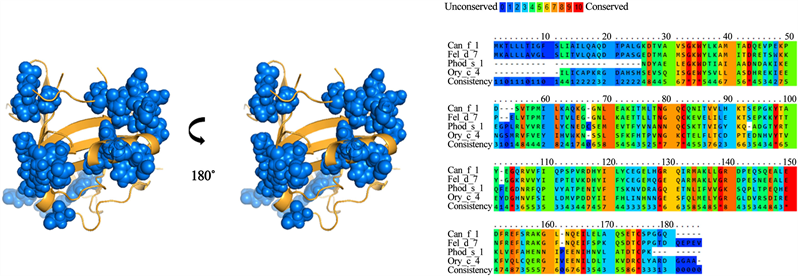
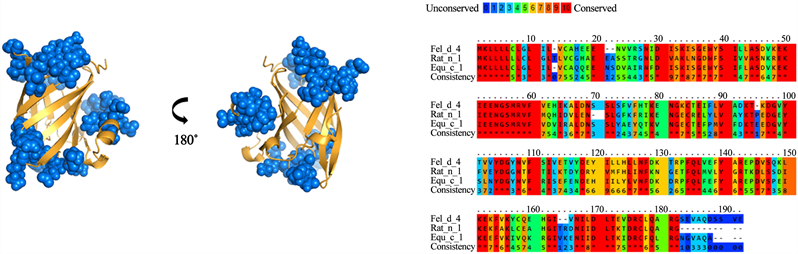
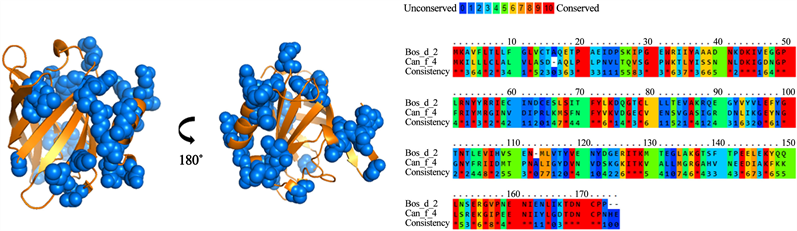
For group B, a 28% identity was found between the amino acid sequences of the lipocalins analyzed (Figure 4). A total of 26 residues were identified and conserved among the different lipocalins from group B. However, when analyzing the antigenic patches, it is observed that they are dispersed in the structure. This suggests that not all the identified residues would be part of the cross-reactivity of these antigens. Meanwhile, group C presented 60% identity in its amino acid sequences (Figure 5). In addition, it presented the highest number of residues exposed and conserved, for a total of 33 residues, which, were concentrated in 3 antigenic patches defined in the lipocalin structure. For groups D and E (Figure 6 and Figure 7), similar results were obtained. The groups shared a 22% and 25% identity between their amino acid sequences, respectively. When analyzing the location of the residues on the structure, it is observed that they present high dispersion and compact antigenic regions are not generated as they are observed in the other groups.
Figure 4. Analysis of group B lipocalins. Blue balls indicate antigenic residues predicted conserved and exposed on surfaces among lipocalins.
Figure 5. Analysis of group C lipocalins. Blue balls indicate antigenic residues predicted conserved and exposed on surfaces among lipocalins.
Figure 6. Analysis of group D lipocalins. Blue balls indicate antigenic residues predicted conserved and exposed on surfaces among lipocalins.
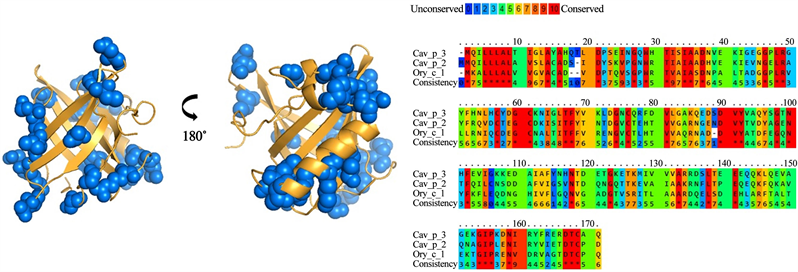
Figure 7. Analysis of group E lipocalins. Blue balls indicate antigenic residues predicted conserved and exposed on surfaces among lipocalins.
4. Discussion
In the present study, we describe potential antigenic regions shared among several lipocalins from domestic animals. Identification of epitopes is crucial to determine the role of cross-reactivity in sensitization to different allergenic sources. Several studies tested cross reactivity in this group of allergens [8] [11] [12] . Can f 1 and Fel d 7 allergens share a 60% of identity in their amino acid sequences and cross reactivity have been demonstrated experimentally [13] . Although both allergens were part of the group B, we found only a 28% of identity between these lipocalins, a low grade of identity to expect cross-reactivity. Our data suggest that cross reactivity of Can f 1, Fel d 7 with Ory c 4 and Phod s 1 is unlikely, due to the identity grade shared between these allergens. Experimental evidence indicates that Phod s 1 allergen is not cross reactive even with extracts from allergenic sources similar to Siberian hamster (Phodopus sungorus) [14] . The closest allergen related to Phod s 1 is Mus m 1, analysis revealed a moderate grade of homology, with a 56% of identity between their amino acid sequences (Data not showed).
Nilsson et al, found cross reactivity between Can f 6, Fel d 4 and Equ c 1, when inhibition assays were performed [12] . In our results, Equ c 1 and Fel d 4 are located in group C, and share phylogenetic relation with Rat n 1, a lipocalin from Rattus novergicus [15] . These lipocalins share a 42% of identity in their amino acid sequences. Our analysis identified 33 residues conserved and surface exposed forming 3 antigenic patches on 3D model. Mostly of antigenic residues identified in C group are conserved in Can f 6, and this lipocalin share a 54% of identity with Rat n 1. This finding suggests that Rat n 1 could be a lipocalin with cross reactivity to Can f 6, Fel d 4 and Equ c 1. Also, we identified potential antigenic regions involved in cross reactivity identified experimentally [15] .
Bos d 2 has been characterized as a weak immunogen. Experimental studies in mice revealed that contains a T cell epitope located in C-terminal region and its amino acid sequence share homology with Can f 1 and Rat n 1 allergens [11] . Bos d 2 bind IgG and IgE antibodies of serum from allergic subjects and induce Th2 proliferative responses in cell lines derived from mice. IgG reacted to C-terminal region of the allergen [11] [16] [17] . This suggests that human and mice recognized to Bos d 2 in a similar way. In our study, we found thirty residues conserved and surface exposed, however, we just find two antigenic regions common between Boss 2 and Can f 1. Maybe, because to the low grade of identity. For E group, Cav p 2, Cav p 3 and Ory c 1 allergen are poorly characterized. These allergens deserve more study because IgE from sixty-five and forty four percent of allergic subjects to guinea pig reacted to Cav p 2 and Cav p 3 [6] . Here, we propose a possible role of Ory c 1 in the cross reactivity with Cav p 2 and Cav p 3, although a low grade of identity was found.
In conclusion, we were able to identify some potential antigenic sites among some lipocalins; however, there is a low identity between these proteins from different species which shows that although cross-reactivity between them is possible, their frequency in most cases is low. These studies support the need to carry out mutagenicity tests to confirm their relevance in the allergenic capacity of lipocalins.
Authors’ Contributions
JS participated in its design and coordination and helped to draft the manuscript. AS and YE participated in the design of the study. MM conceived of the study and performed in silico analysis. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no conflicts of interest regarding the publication of this paper.
Cite this paper
Marlon, M., Andres, S., Jorge, S. and Yuliana, E. (2018) In Silico Analysis of Cross Reactivity between Lipocalin of Domestic Animals. Open Journal of Immunology, 8, 97-106. https://doi.org/10.4236/oji.2018.84006
References
- 1. Simons, F.E.R., Ardusso, L.R.F., Bilò, M.B., El-Gamal, Y.M., Ledford, D.K., Ring, J., et al. (2011) World Allergy Organization Guidelines for the Assessment and Management of Anaphylaxis—The 2017 Revision and Update. WAOJ, 4, 13-37. https://doi.org/10.1097/WOX.0b013e318211496c
- 2. Scheurer, S., Toda, M. and Vieths, S. (2015) What Makes an Allergen? Clinical & Experimental Allergy, 45, 1150-1161. https://doi.org/10.1111/cea.12571
- 3. Karp, C.L. (2010) Guilt by Intimate Association: What Makes an Allergen an Allergen? Journal of Allergy and Clinical Immunology, 125, 955-960. https://doi.org/10.1016/j.jaci.2010.03.002
- 4. Virtanen, T. (2001) Lipocalin Allergens. Allergy, 56, 48-51. https://doi.org/10.1034/j.1398-9995.2001.00915.x
- 5. Virtanen, T., Zeiler, T. and Mantyjarvi, R. (1999) Important Animal Allergens Are Lipocalin Proteins: Why Are They Allergenic? International Archives of Allergy and Immunology, 120, 247-258. https://doi.org/10.1159/000024277
- 6. Hilger, C., Swiontek, K., Kler, S., Diederich, C., Lehners, C., Vogel, L., et al. (2011) Evaluation of Two New Recombinant Guinea-Pig Lipocalins, Cav p 2 and Cav p 3, in the Diagnosis of Guinea-Pig Allergy. Clinical & Experimental Allergy, 41, 899-908. https://doi.org/10.1111/j.1365-2222.2011.03726.x
- 7. Ganfornina, M.D., Gutierrez, G., Bastiani, M. and Sanchez, D. (2000) A Phylogenetic Analysis of the Lipocalin Protein Family. Molecular Biology and Evolution, 17, 114-126. https://doi.org/10.1093/oxfordjournals.molbev.a026224
- 8. Andrés Sánchez, R.C. and Sánchez, J. (2017) Analysis in Silico of Lipocalinas of Dog, Cat, Horse, Cow, Hamster and Hen. Possible Effect in the Study of Allergic Diseases. Rev Alerg Méx, 64.
- 9. Jennifer, B., Nuttall, T.J., Hammerberg, B. and Halliwell, R.E. (2017) Co-Sensitization and Cross-Reactivity between Related and Unrelated Food Allergens in Dogs—A Serological Study. Veterinary Dermatology, 28, 31-e7. https://doi.org/10.1111/vde.12335
- 10. Simossis, V.A. and Heringa, J. (2005) PRALINE: A Multiple Sequence Alignment Toolbox That Integrates Homology-Extended and Secondary Structure Information. Nucleic Acids Research, 33, 89-94.
- 11. Immonen, A., Farci, S., Taivainen, A., Partanen, J., Pouvelle-Moratille, S., Närvänen, A., et al. (2005) T Cell Epitope-Containing Peptides of the Major Dog Allergen Can f 1 as Candidates for Allergen Immunotherapy. The Journal of Immunology, 175, 3614-3620. https://doi.org/10.4049/jimmunol.175.6.3614
- 12. Nilsson, O.B., Binnmyr, J., Zoltowska, A., Saarne, T., van Hage, M. and Gronlund, H. (2012) Characterization of the Dog Lipocalin Allergen Can f 6: The Role in Cross-Reactivity with Cat and Horse. Allergy, 67, 751-757. https://doi.org/10.1111/j.1398-9995.2012.02826.x
- 13. Apostolovic, D., Sanchez-Vidaurre, S., Waden, K., Curin, M., Grundstrom, J., Gafvelin, G., et al. (2016) The Cat Lipocalin Fel d 7 and Its Cross-Reactivity with the Dog Lipocalin Can f 1. Allergy, 71, 1490-1495. https://doi.org/10.1111/all.12955
- 14. Torres, J.A., de Las Heras, M., Maroto, A.S., Vivanco, F., Sastre, J. and Pastor-Vargas, C. (2014) Molecular and Immunological Characterization of the First Allergenic Lipocalin in Hamster: The Major Allergen from Siberian Hamster (Phodopus sungorus). The Journal of Biological Chemistry, 289, 23382-23388. https://doi.org/10.1074/jbc.M114.579060
- 15. Zeiler, T., Mantyjarvi, R., Rautiainen, J., Rytkonen-Nissinen, M., Vilja, P., Taivainen, A., et al. (1999) T Cell Epitopes of a Lipocalin Allergen Colocalize with the Conserved Regions of the Molecule. The Journal of Immunology, 162, 1415-1422.
- 16. Saarelainen, S., Zeiler, T., Rautiainen, J., Narvanen, A., Rytkonen-Nissinen, M., Mantyjarvi, R., et al. (2002) Lipocalin Allergen Bos d 2 Is a Weak Immunogen. International Immunology, 14, 401-409. https://doi.org/10.1093/intimm/14.4.401
- 17. Saarelainen, S.A., Kinnunen, T.T., Buhot, C., Narvanen, A.T., Kauppinen, A.K., Rytkonen-Nissinen, M.A., et al. (2008) Immunotherapeutic Potential of the Immunodominant T-Cell Epitope of Lipocalin Allergen Bos d 2 and Its Analogues. Immunology, 123, 358-366. https://doi.org/10.1111/j.1365-2567.2007.02699.x
NOTES
*Lipocalins bioinformatics analysis.