Predicting Yield and Stability Analysis of Wheat under Different Crop Management Systems across Agro-Ecosystems in India ()
1. Introduction
Worldwide, wheat is the most important staple food as it provides 55% of the carbohydrates and 20% of the food calories and protein consumed globally [1] . Demand for wheat is projected to continue to grow over the coming decades, particularly in the developing world to feed an increasing population, and with wheat being a preferred food, continuing to account for a substantial share of human energy needs in 2050 [2] . In India, wheat is the second most important cereal crop after rice. During the past century, India has achieved remarkable progress in increasing wheat production and productivity, primarily due to genetically improved cultivars, expansion of sown areas and better cultural methods. Improved agronomic practices played an important role in enhancing the dependability and sustainability of yields [3] [4] . Yield is a complex quantitative trait and greatly influenced by external environment, which results in scale or rank shift in its performance [5] [6] . This relative shift of genotype performance from one environment (location x year combination) to another is known as genotype x environment interaction (GxE) [7] [8] . Substantial studies have been conducted to identify high yielding and consistent performing wheat genotypes (also known as stable genotypes). However, most of the high stable genotypes are less predictable across different crop management practices since plant breeders often perform analysis of two-way data (genotype x site or GxE) for several consecutive years to detect stable genotypes without taking crop management practices into account. Previous studies on tillage practices, a crop management practice, suggest that optimization of tillage practice alters the dynamic and complex soil in physical, chemical and biological properties [9] - [15] . Proponents of conventional tillage (CT) and zero tillage (ZT) agree on the values of one over the other for crop yield variation across locations and cropping systems [16] - [21] . Understanding interactions among genotype, environment and crop management are important in planning key farm decisions including genotype selection, sustainable farm management and economic planning. In this paper, we propose to identify high yielding and high stable wheat genotypes across environments and over two-crop management practices: CT and ZT.
The impenetrable interaction of a crop bio-system with the soil, the atmosphere, and the environment that a plant lives in introduces challenges when making breeding decisions because it complicates the demonstration of superiority of any genotype across environments. Genotype x environment interaction may result in low correlation between phenotypic and genotypic values, thereby reducing progress from selection. This reduction leads to bias in the estimation of heritability and in the prediction of genetic advance [22] [23] [24] . Several strategies have been proposed to deal with GxE, and the most powerful strategy is to exploit GxE either to develop locally adapted material or to use GxE to better characterize the genotypes [25] [26] . Genotype x environment interaction can be characterized using statistical methods ranging from univariate to multivariate models. The univariate models include regression slope, deviation from regression, environmental variance, and Kang’s yield-stability; multivariate models include genotype main effects plus genotype by environment interaction (GGE) biplot, and additive main effects and multiplicative interaction (AMMI) [27] - [33] . Multivariate models could be graphically represented through biplots where genotypes and environments are plotted in a single graph. Recently, hierarchical Bayesian and mixed models were introduced to model heterogeneous variance among environments and different correlation structures among environments [26] [34] [35] [36] [37] . Mixed models allow more flexibility to model unbalanced data using restricted maximum likelihood estimates (REML). Each statistical method reflects different aspects of the GxE, and no single method adequately explains genotype performance across environments [6] . Stability statistics are best used in combination with trait performance (mean or BLUP: Best Linear Unbiased Predictor is an estimate of random effect) and have successfully been used in plant breeding.
In this study, we were interested in understanding differential sensitivity of certain wheat genotypes to different agro-ecological environments in India under different crop management practices for enhancing the selection of superior and stable genotypes. The objectives of the study were as follows: 1) to evaluate the genotype x environment x management interaction (GxExM) for wheat genotypes; 2) to predict yield performance and identify high stable wheat genotypes in different management practices; and 3) to make genotype-specific management and high performing genotype recommendations within and across agro-ecological regions.
2. Materials and Methods
2.1. Germplasm, Location and Management
Twenty-one genotypes of wheat were evaluated across three years (2012, 2103, and 2014) and three locations ranging from western to eastern Indo-Gangetic plains under two different tillage management practices. Locations were chosen to represent the major wheat growing agro-ecological conditions for major wheat production regions in India: Pusa (25˚57'08''N; 85˚40'13''E), Bihar (BR), Jabalpur (23˚10'7.6''N; 79˚55'55''E), Madhya Pradesh (MP), and Ludhiana (30˚59'28''N; 75˚44'11''E), Punjab (PB) (Supplemental Figure S1 and Supplemental Figure S2). The soils of the experimental plots at three locations, PB, MP and BR, were sandy loam, clay loam and silty loam, respectively. Twenty-one genotypes were semi-dwarf spring wheat chosen to represent new vs. old release; stress resistant vs. susceptible; rust resistant vs. susceptible; eastern vs. northern adapted; tall vs. short stature; early vs. late maturity; low vs. high yield; and varied 1000 kernel weight, protein content, spikelet ear−1 and seeds spike−1. These genotypes are frequently used advanced breeding lines or officially released cultivars. Hereafter the word “genotype” is used to indicate cultigen, cultivar, variety or genotype. Genotypes were considered to be random and representative samples of a wide range of genetic and phenotypic diversity in the wheat germplasm population (Supplemental Table S1). Wheat genotypes were evaluated at two levels of crop management, a conventional tillage (CT) and zero tillage (ZT). The CT involved two plowings with disc harrow, the first plowing used a spring-tyne cultivator (about 0.15 m depth) followed by planking at optimum soil moisture content; and the second plowing involved seeding of different varieties using “Limit-Plot Planter”. In ZT, different varieties were directly drilled without any preparatory tillage operations and in the presence of standing stubbles of rice residues (~15 cm) using “Limit-Plot Planter” with inverted “T” tyne openers [38] [39] . A standard seed rate (100 kg∙ha−1), seeding depth (4 cm), fertilizer nutrient application (120 kg N, 60 kg P2O5 and 60 kg K2O ha−1), water management (irrigation at standard critical stages) was adopted for both CT and ZT. These tillage and other management practices correspond to the crop management system commonly used in commercial wheat production in India [40] [41] .
2.2. Trials and Data Description
At each location, individual trials were established as a two-factor strip-plot (split-block) design with ten genotypes, three replications and two-crop management practices [42] . Out of the ten genotypes, six genotypes were tried at all three locations for three years, and the remaining four genotypes were location- specific and not necessarily duplicates. These four location-specific genotypes were evaluated at one or two location(s) for either one or all three years. Thus, total distinct genotypes were twenty-one (Supplemental Table S1). Within the blocks, the genotypes were arranged in sub-blocks, and the two-crop managements in the other sub-blocks were arranged perpendicularly to the sub-blocks with the genotypes. The experimental unit (harvest plot) size was 1.6 m × 10 m.
In each location and year, the ten wheat genotypes were evaluated for grain yield (t∙ha−1). Wheat grains were harvested using the guide of15% moisture in grain, yellowing of spikelets and hard dough stage of grains [11] . Data were missing at random from one replication, one location and one year for genotypes “BAZ”, “CSW-16”, “CSW16”, “CSW-18”, and “DPW-621-50”. We excluded three genotypes (“GW-273”, “GW-366” and “HD-2687”) that were tested for a time period shorter than three years and only in one location to provide a sufficient representative sample of years and location as random and fixed factors, respectively. As a result of this data preparation, we obtained a four-way genotype x management x location x year interaction (GxMxLxY) dataset for the grain yield of eighteen wheat genotypes (Supplemental Table S1). A set of six genotypes evaluated in all locations were analyzed for across-location statistics. Similarly, a set of ten genotypes (six genotypes from all locations + four unique genotypes from each location) were analyzed for individual location statistics (Supplemental Table S1).
2.3. Data Analysis and Statistical Methods
The unbalanced grain yield GxMxLxY data were analyzed for genotype, environment, management, and genotype x environment x management interactions with the SASGxE [6] [43] [44] and RGxE [45] [46] programs using SAS and R programming language, respectively, in two steps. During the first stage, we imputed the missing values using the mice() function of the mice (multivariate imputation by chained equations) package [47] of R. Parameters maximum iteration 50, predictive mean matching (pmm) method and random generator seed value 500 were used in the mice() function to generate five imputed datasets. Then, in the second stage, the five imputed datasets were combined across trials and years to obtain a balanced GxMxLxY mean data for all statistical analyses.
Years and genotypes, and locations and managements were analyzed as random and fixed effects, respectively. Estimates and significance of random effects were computed using RGxE. The random effect model was fit using the lmer() function of lme4 (linear mixed effects models) package [48] . The F ratio (= MSbetween/MSwithin, where MS is mean square or variance estimate) and significance of fixed effects were computed using mixed() function of afex package [49] . The mixed() function computes type III such as p-values using the default method via the Kenward-Roger approximation for degrees of freedom. Similarly, the significance of random effects was computed using a likelihood ratio test to attain p-values. Likelihood is the probability of the data given a model. The logic of the likelihood ratio test is to compare the likelihood of two models with each other using restricted maximum likelihood (REML) methodology. The model without the factor of interest (the null model) is compared with the model with the factor of interest (the full model) using the anova() function. It gives a Chi- Square value, the associated degrees of freedom and p-value. According to Wilk’s theorem, the negative two times the log likelihood ratio of two models approaches a Chi-Square distribution with k degrees of freedom, where k is number of random effects tested [50] .
Univariate stability statistics [regression slope (bi), deviation from regression (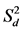 ), Shukla’s stability variance (
), Shukla’s stability variance (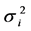 ), and Kang’s yield-stability statistics (YSi)], and BLUP for genotypes were computed using RGxE. Regression slope (bi) and deviation from regression (
), and Kang’s yield-stability statistics (YSi)], and BLUP for genotypes were computed using RGxE. Regression slope (bi) and deviation from regression (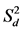 ); Shukla’s stability variance (
); Shukla’s stability variance ( ) and Kang’s yield-stability statistics (YSi); and best linear unbiased predictor (BLUP) for genotypes were computed using lm() function of R [51] stability.par() function of the agricolae package [52] and ranef() function of lme4 package [48] , respectively. Tests for significance were derived using a t-test for each bi and an F test for each
) and Kang’s yield-stability statistics (YSi); and best linear unbiased predictor (BLUP) for genotypes were computed using lm() function of R [51] stability.par() function of the agricolae package [52] and ranef() function of lme4 package [48] , respectively. Tests for significance were derived using a t-test for each bi and an F test for each 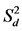 for statistical differences from one and zero, respectively, at 0.05, 0.01 and 0.001 levels of probability.
for statistical differences from one and zero, respectively, at 0.05, 0.01 and 0.001 levels of probability.
SASGxE provided R code that is ready to use in R statistical software [51] for the analysis of multivariate stability statistics (GGE biplot) [44] . GGE biplot analysis was computed using the “GGEBiplotGUI” package [53] , with the support in the helper application “RStudio” [54] in R statistical software. GGE biplot analysis was used to visually assess the presence of genotype x environment interaction and rank genotype based on stability and mean in each management practice [32] [55] . For each management practice, input data of GGE biplot analysis consisted of genotype x environment matrix (2 × 2) of mean values.
Similarly performing genotypes across and within locations and years were clustered using PROC VARCLUS of SAS v9.4 [56] . The VARLCUS procedure used user-defined second eigenvalue cutoff and underlying algorithm called divisive clustering to split a given set of genotypes into two groups. Eigenvalues are the coefficients of principal component analysis. The value 1 of the second eigenvalue is a common choice for cut off because it represents the average size of the eigenvalues. However, we have used the smaller value of the second eigenvalue as 0.7 to account for sampling variability [57] . PROC VARCLUS identified clusters and computed 1 − R2 ratio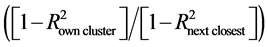 , which identifies a cluster of genotypes that are highly correlated among themselves and not highly correlated with genotypes in other clusters. The graphical representation of divisive clustering, 1 − R2 ratio, forest plot of BLUP along with prediction interval, and the stability statistics summary of each genotype in different management practices across and within locations were computed using SAS PROC TEMPLATE in conjugation with PROC SGRENDER while utilizing the graphical template language (GTL) of SAS v9.4.
, which identifies a cluster of genotypes that are highly correlated among themselves and not highly correlated with genotypes in other clusters. The graphical representation of divisive clustering, 1 − R2 ratio, forest plot of BLUP along with prediction interval, and the stability statistics summary of each genotype in different management practices across and within locations were computed using SAS PROC TEMPLATE in conjugation with PROC SGRENDER while utilizing the graphical template language (GTL) of SAS v9.4.
3. Results
3.1. Variance Analysis
The pooled analysis revealed statistically significant fixed effects (L, M, MxL) for grain yield (Table 1). The variance estimates of the yield for the year (Y) and the
![]()
Table 1. Pooled variance analysis for wheat yield (t ha−1) of 6 genotypes tested in 3 years and 3 locations over 2 management practices; and location variance of 10 genotypes tested for 3 years over 2 management practices.
*, **, and *** = significant at 0.05, 0.01, and 0.001 levels of probability, respectively; NS =non-significant. The value of significance presented in “F-ratio” column represents F statistics p-values for F-ratio of fixed effects. Similarly, the value of significance presented in “% of total variance” column represents χ2 statistics p-values of variance estimate of random effects. †Degrees of freedom: Y-2; L-2, YxL-4; G-5; GxY-10; GxL-10; GxYxL-20; M-1; MxY-2; MxL-2; MxLxY-4; GxM-5; GxMxY-10; GxMxL-10; GxMxLxY-20. ‡Degrees of freedom: Y-2; G-9; GxY-18; M-1; MxY-2; GxM-9; GxMxY-18. ¶% total variance is calculated by variance estimate of given factor divided by sum (total) of variance estimate of all the factors. List of factors used in model are presented in source column.
interaction between year and location (YxL) were different (p < 0.01) from zero and accounted for 39% of the total variation. Except for GxY and MxY, estimates of the random effects were significant (ranged from 0.17% - 24% of the total variance estimate). Large estimate of environment (LxY) variance dominated the expression of genotype effect (G) and interaction between genotype and management (GxM) (Table 1). Within location analysis, both fixed and random effects were significant in BR, MP and PB. In BR and MP, Y, G and GxM caused most of the variation in yield performance of wheat genotype (22% and 39%, 35% and 24%, and 26% and 21% of the total variance, respectively). In contrast, PB had large Y effect (53% of total variance), medium G effect (12% of total variation) and small GxM effect (3% of total variation) (Table 1).
3.2. Polygon View of GGE Biplot
The “polygon” (or which-won-where) view of the GGE biplot is drawn by joining extreme genotypes and perpendicular lines (rays) passing from the polygon sides divides the biplot into sector (Figure 1 and Figure 2). A crossover GxE pattern exists if environments fall into multiple sectors and, thus, different genotypes win for an environment or set of environments in a sector. Conversely, no GxE pattern exists if all environments fall into a single sector and, thus, a single genotype had the highest yield in all environments. In either case, the winning genotype is vertex genotype. If no environment is present in a sector, then vertex genotype of that sector is considered to be a poor performer in all test environments. Genotypes within the polygon were less responsive to location than the vertex genotypes. Across locations, the polygon view of the GGE biplot explained 96% and 97% of the genotype and genotype x environment variation for the management practice CT and ZT, respectively (Figure 1: Panel A and B). For management practice CT, environments were grouped into two sectors with different winning (vertex) genotypes. This confirms the existence of GxE for CT. Conversely, ZT had all the locations grouped into one sector, which suggests that GxE for ZT did not exist. However, the GGL biplots for individual year confirm that the location grouping varied across years for ZT. Results of GGL are presented in Supplemental Figure S3 (Panel B, Panel D and Panel E).
Within location, a polygon view of the GGY biplot explained 96% and 98%, 88% and 84%, and 96% and 95% for the management practice CT and ZT, and location BR, MP and PB, respectively (Figure 2: Panel A and Panel B; Panel C and Panel D; Panel E and Panel F). For location BR, environments grouped into two and one sector(s) for management practice CT and ZT, respectively (Figure 2: Panel A and Panel B). This suggests that management practice ZT had single winning genotypes across years, whereas CT had different winning genotypes across years. Location MP had environments grouped into two sectors with different winning (vertex) genotypes in each management practice (Figure 2: Panel C and Panel D). Similarly, for location PB and both management practices, environments were grouped into two sectors with different winning genotypes. However, the pattern of winning genotype and environment grouping are con-
![]()
Figure 1. The polygon (which-won-where) (Panel A and Panel B) and mean vs. stability (Panel C and Panel D) view of genotype main effects plus genotype x environment interaction effect (GGE) biplot of yield of 6 wheat genotypes tested in 3 years, 3 locations over 2 management practices. The biplots were based on Scaling = 0, Centering = 0, and SVP = 2. Key to the labels of genotype and location is presented in abbreviation section.
sistent in year 2012 (Figure 2: Panel E and Panel F).
3.3. Genotype BLUPs
BLUPs are the estimates of random effects. Across locations, estimates of genotype (random effect) for wheat yield ranged from 5.03 to 5.55 t∙ha−1 and 5.01 to 5.64 t∙ha−1 for management practice CT and ZT, respectively (Table 2). For both management practices (CT and ZT), the highest yield was estimated for genotype “HD-2967” and the lowest yield was estimated for genotype “CSW-18”. Other high yielding genotypes were “Munal”, “BAZ” and “CSW-16”.
Within individual location under both management practices, the predicted yield ranged from 4.67 to 5.87 t∙ha−1 and 4.64 to 6.09 t∙ha−1; 4.84 to 5.17 t∙ha−1 and 4.77 to 5.14 t∙ha−1; and 5.06 to 5.50 t∙ha−1 and 5.03 to 5.57 t∙ha−1 for location BR, MP and PB, and management practice CT and ZT, respectively (Table 2). Under both management practices, location BR had genotypes “HD-2967” and “CSW-16” with the highest and lowest predicted yield, respectively. For location MP, the highest yield was predicted for genotype “GW-322” and “MPO-1215” in management practice CT and ZT, respectively. Genotypes “JW-3288” and “HD- 2932” had the lowest predicted yield for management practice CT and ZT, respectively. Location PB had the highest predicted yield for “Munal” and “HD-
![]()
![]()
Figure 2. The polygon (which-won-where) view of genotype main effects plus genotype x year interaction effect (GGY) biplot of location BR (Panel A and Panel B), MP (Panel C and Panel D) and PB (Panel E and Panel F) for yield of 10 (6 same + 4 different) wheat genotypes tested in 3 years over 2 management practices. The biplots were based on Scaling = 0, Centering = 0, and SVP = 2. Key to the labels of genotype and location is presented in abbreviation section.
2967” in CT and ZT, respectively, and the lowest predicted yield for “CSW-16” in both management practices.
3.4. Univariate Stability Statistics
According to Eberhart and Russell (1966), a regression coefficient (bi) approximating unity, along with deviation from regression ( ) near zero, indicates stability. Across locations, the bi value for all six genotypes was close (p > 0.001) to unity under both management practices, except for “DPW-621-50” in ZT (Supplemental Table S2). Conversely, the
) near zero, indicates stability. Across locations, the bi value for all six genotypes was close (p > 0.001) to unity under both management practices, except for “DPW-621-50” in ZT (Supplemental Table S2). Conversely, the 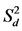 value of all six evaluated genotypes was significantly different (p < 0.05) from zero, except for “BAZ” in CT. Within individual location under both management practices bi and
value of all six evaluated genotypes was significantly different (p < 0.05) from zero, except for “BAZ” in CT. Within individual location under both management practices bi and  values were close to unity (p > 0.001) and zero (p > 0.001), respectively, for all ten tested genotypes. However, the exceptions were “Munal” and “JW-3288” in location MP under management practice CT for bi and
values were close to unity (p > 0.001) and zero (p > 0.001), respectively, for all ten tested genotypes. However, the exceptions were “Munal” and “JW-3288” in location MP under management practice CT for bi and 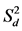 values significantly different from one and zero, respectively, and “CSW-16” in location BR under management practice ZT for
values significantly different from one and zero, respectively, and “CSW-16” in location BR under management practice ZT for 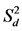 value significantly different zero. For management practice CT, genotypes “HD-2967”; “HD-2932”, “HD-2967” and “JW-3288”; and “DBW-88” had a negative bi value in location BR, MP, and PB, respectively. Similarly, genotype “MPO-1215” had a negative bi value in location MP under management practice ZT.
value significantly different zero. For management practice CT, genotypes “HD-2967”; “HD-2932”, “HD-2967” and “JW-3288”; and “DBW-88” had a negative bi value in location BR, MP, and PB, respectively. Similarly, genotype “MPO-1215” had a negative bi value in location MP under management practice ZT.
According to Shukla (1972), a genotype with low  is regarded as stable. Across locations, “BAZ” and “DPW-621-50” had significantly low
is regarded as stable. Across locations, “BAZ” and “DPW-621-50” had significantly low  in management practices CT and ZT (Supplemental Table S2). Similarly, under both management practices, genotypes “DPW-621-50”; “BAZ” and “DPW-621-50”; and “DPW-621-50”, “HD-2967”, “Munal”, “PBW-343” and “PBW-550” had low
in management practices CT and ZT (Supplemental Table S2). Similarly, under both management practices, genotypes “DPW-621-50”; “BAZ” and “DPW-621-50”; and “DPW-621-50”, “HD-2967”, “Munal”, “PBW-343” and “PBW-550” had low 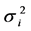 in location BR, MP, and PB, respectively.
in location BR, MP, and PB, respectively.
According to YSi, genotypes with YSi higher than the mean YSi are stable (represented with symbol “√” in Supplemental Table S2). Across locations, genotypes “BAZ”, “DPW-621-50”, “HD-2967” and “Munal” were stable in both management practices. For location BR, stable genotypes in both management practices were “CSW-16”, “DPW-621-50”, “HD-2733”, “HD-2824”, and “HD-2967”. Similarly, genotypes “BAZ”, “GW-322”, “HD-2967”, “MPO-1215”, and “Munal” were stable in MP across both management practices. Likewise, in location PB genotypes “DPW-621-50”, “HD-2967”, “Munal”, and “PBW-550” were stable in both management practices.
3.5. Mean vs. Stability, View of GGE Biplot
The “average environment coordinate” (AEC) view based on genotype-focused singular value partitioning (SVP = 1) can be referred to as the “mean vs. stability” view [58] of GGE biplot. That view facilitates genotype comparisons based on mean performance and stability across environments within a mega-environ- ment. The “mean vs. stability” view of GGE biplot explained 96% and 99% of genotypic and genotype x environment variation across locations under management practices CT and ZT, respectively (Figure 1: Panel C and D). The arrow shown on the AEC abscissa points in the direction of higher yield performance of genotypes and ranks the genotypes with respect to yield performance. Thus, genotype “HD-2967” (G14) had the highest yield and “CSW-18” (G03) had the lowest under both management practices (Figure 1: Panel C and D). The stability of each genotype was explored by its projection onto the AEC vertical axis. The most stable genotype was located almost on the AEC abscissa (horizontal axis) and had a near-zero projection onto the AEC (vertical axis). Thus, “HD- 2967” (G14) and “DPW-621-50” (G05), and “Munal” (G19) and “BAZ” (G01) were the most stable in management practices CT and ZT, respectively (Figure 1: Panel C and D). The ideal genotype has both the high trait mean and stable performance [46] . An ideal genotype is represented by a circle on the head of arrow on the AEC abscissa (horizontal axis) (Figure 1: Panel C and D). Across locations, genotype “HD-2967” (G14) was the best in both management practices (Figure 1: Panel C and D). These findings are consistent across years (Supplemental Figure S4).
For location BR, most of the genotypes were clustered near the origin (0,0) of the biplot, which represents the average yield. In CT, “K-307” (G17), “CSW-16” (G02), “HD-2824” (G12) and “HD-2967” (G14) were best. Similarly, in ZT. “BAZ” (G01), “HD-2733” (G11) and “DPW-621-50” (G05) were best (Figure 3: Panel A
![]()
Figure 3. The mean vs. stability view of genotype main effects plus genotype x year interaction effect (GGY) biplot of location BR (Panel A and Panel B), MP (Panel C and Panel D) and PB (Panel E and Panel F) for yield of 10 (6 same + 4 different) wheat genotypes tested in 3 years over 2 management practices. The biplots were based on Scaling = 0, Centering = 0, and SVP = 2. Key to the labels of genotype and location is presented in abbreviation section.
and Panel B). For location MP, “Munal” (G19) and “MPO-1215” (G18), and “GW-322” (G08) were best in CT and ZT, respectively (Figure 3: Panel C and Panel D). For location PB, “HD-2967” (G14), and “PBW-550” (G21) were best in both management practices (Figure 3: Panel E and Panel F).
3.6. Divisive Clusters and 1 − R2 ratio
Across locations under both management practices, similarly performing genotypes were grouped into two clusters (Figure 4: Panel A and Panel B). The most representative and distinct genotype within the cluster has high correlation with its own cluster and low correlation with other clusters [59] . Thus, an ideal representative genotype has a low 1 − R2 ratio ![]() value. Across all the tested locations “Munal” and “HD-2967” were the representative genotype of cluster 1 and cluster 2, respectively, in CT (Figure 4: Panel A).
value. Across all the tested locations “Munal” and “HD-2967” were the representative genotype of cluster 1 and cluster 2, respectively, in CT (Figure 4: Panel A).
![]()
Figure 4. Divisive cluster, 1 − R2 ratio, forest plot of BLUP along with 95% prediction interval, and stability statistics summary of yield of 6 wheat genotypes tested in 3 years, 3 locations over 2 management practices. † *, ** and *** significantly different from unity for the regression coefficients or slope (bi) and from zero for the deviation from regression (![]() ) and Shukla’s stability variance (
) and Shukla’s stability variance (![]() or Si) at 0.05, 0.01 and 0.001 levels of probability, respectively. NS is significantly not different. “§” indicates negative slope. “+” and “−” indicate stable and non-stable according to Kang stability statistics (YSi), respectively. “HYHS”, “HYLS”, “MYHS”, “MYLS”, “LYHS” and “LYLS” indicate “high yield and high stable”, “high yield and low stable”, “medium yield and high stable”, “medium yield and low stable”, “low yield and high stable” and “low yield and low stable”, respectively. High, medium and low yield is with reference to the center of “mean vs. stability” view of GGE biplot (0,0), which represents the mean yield.
or Si) at 0.05, 0.01 and 0.001 levels of probability, respectively. NS is significantly not different. “§” indicates negative slope. “+” and “−” indicate stable and non-stable according to Kang stability statistics (YSi), respectively. “HYHS”, “HYLS”, “MYHS”, “MYLS”, “LYHS” and “LYLS” indicate “high yield and high stable”, “high yield and low stable”, “medium yield and high stable”, “medium yield and low stable”, “low yield and high stable” and “low yield and low stable”, respectively. High, medium and low yield is with reference to the center of “mean vs. stability” view of GGE biplot (0,0), which represents the mean yield.
Similarly, “BAZ” was the representative genotype of cluster 1 in ZT (Figure 4: Panel B). The zero value of 1 − R2 ratio is the result of the presence of a single entity in the cluster and, thus, correlation within its own cluster is 1 ![]() (Figure 4: Panel B).
(Figure 4: Panel B).
For location BR, similarly performing genotypes across years were grouped into three clusters under both management practices (Figure 5: Panel A and Panel B). In CT, “HD-2824” and “DPW-621-50” were representative genotypes in cluster 1 and cluster 3, respectively (Figure 5: Panel A). In ZT, “Munal”, “CSW-16” and “HD-2967” were representative in respective clusters (Figure 5: Panel B). Similarly, for location MP, 5 and 2 clusters were identified in CT and ZT, respectively (Figure 5: Panel C and Panel D). In CT, “HD-2932”, “BAZ” and “JW-3288” were representative in cluster 1, cluster 3 and cluster 4, respectively (Figure 5: Panel C). In ZT, “GW-322” and “Munal” were representative in cluster 1 and cluster 2, respectively (Figure 5: Panel D). For location PB under both management practices, genotypes were grouped into 1 cluster, except for “DBW- 88” in CT (Figure 6: Panel A and B).
4. Discussion
Across locations, the F ratio of the fixed effects (L, M, MxL) was statistically significant. The mean yield of genotypes for the crop management practices (M), CT and ZT, was different (results are not presented here) and these results are in agreement with our previous findings [11] [12] [18] [19] [20] [21] [60] [61] . The significant MxL suggests a variable response of genotype yield to the increased intensity of tillage practices across the test locations. Except for GxY and MxY, estimates of the random effects were significant (Table 1). Similar results were reported in the study by [62] [63] [64] [65] [66] . The estimates of random effects varied from 0.17% - 24% of the total variance. The large estimates of Y and YxL suggest that the agro-ecological conditions of the test locations were extremely different and accounted for most of the yield variation. Variations in meteorological data across locations confirm the diversity found in agro-ecological conditions of the test locations (Supplemental Figure S1 and Supplemental Figure S2). The small contribution of G in the total variance estimate is due to the fact that genotypes evaluated in this study were advanced breeding lines, elite cultivars or high yielding genotypes. These findings are confirmed with the common parents and number of selfing (inbreeding) found in their pedigree (Supplemental Table S1). The significant variance components for GxM, GxMxL and GxMxLxY led to a different ranking of genotypes across environments under CT and ZT, justifying the development of a stable genotype that performs well over environments in different management practices. The ideal genotype should have a high mean and high stability.
Across locations, the mean predicted (mean of BLUP) yield of ZT is more than CT (represented by the vertical line in Figure 4: Panel A and Panel B). A similar trend of enhanced performance of ZT over CT across nine environments (three locations × three years) was noticed (Supplemental Figure S5). This
![]()
Figure 5. Divisive cluster, 1 − R2 ratio, forest plot of BLUP along with 95% prediction interval, and stability statistics summary of locations BR (Panel A and Panel B) and MP (Panel C and Panel D) for yield of 10 (6 same + 4 different) wheat genotypes tested for 3 years over 2 management practices. † *, ** and *** significantly different from unity for the regression coefficients or slope (bi) and from zero for the deviation from regression (![]() ) and Shukla’s stability variance (
) and Shukla’s stability variance (![]() or Si) at 0.05, 0.01 and 0.001 levels of probability, respectively. NS is significantly not different. “§” indicates negative slope. “+” and “−” indicate stable and non-stable according to Kang stability statistics (YSi), respectively. “HYHS”, “HYLS”, “MYHS”, “MYLS”, “LYHS” and “LYLS” indicate “high yield and high stable”, “high yield and low stable”, “medium yield and high stable”, “medium yield and low stable”, “low yield and high stable” and “low yield and low stable”, respectively. High, medium and low yield is with reference to the center of “mean vs. stability” view of GGE biplot (0,0), which represents the mean yield.
or Si) at 0.05, 0.01 and 0.001 levels of probability, respectively. NS is significantly not different. “§” indicates negative slope. “+” and “−” indicate stable and non-stable according to Kang stability statistics (YSi), respectively. “HYHS”, “HYLS”, “MYHS”, “MYLS”, “LYHS” and “LYLS” indicate “high yield and high stable”, “high yield and low stable”, “medium yield and high stable”, “medium yield and low stable”, “low yield and high stable” and “low yield and low stable”, respectively. High, medium and low yield is with reference to the center of “mean vs. stability” view of GGE biplot (0,0), which represents the mean yield.
![]()
Figure 6. Divisive cluster, 1 − R2 ratio, forest plot of BLUP along with 95% prediction interval, and stability statistics summary of locations PB for yield of 10 (6 same + 4 different) wheat genotypes tested for 3 years over 2 management practices. † *, ** and *** significantly different from unity for the regression coefficients or slope (bi) and from zero for the deviation from regression (![]() ) and Shukla’s stability variance (
) and Shukla’s stability variance (![]() or Si) at 0.05, 0.01 and 0.001 levels of probability, respectively. NS is significantly not different. “§” indicates negative slope. “+” and “-” indicate stable and non-stable according to Kang stability statistics (YSi), respectively. “HYHS”, “HYLS”, “MYHS”, “MYLS”, “LYHS” and “LYLS” indicate “high yield and high stable”, “high yield and low stable”, “medium yield and high stable”, “medium yield and low stable”, “low yield and high stable” and “low yield and low stable”, respectively. High, medium and low yield is with reference to the center of “mean vs. stability” view of GGE biplot (0,0), which represents the mean yield.
or Si) at 0.05, 0.01 and 0.001 levels of probability, respectively. NS is significantly not different. “§” indicates negative slope. “+” and “-” indicate stable and non-stable according to Kang stability statistics (YSi), respectively. “HYHS”, “HYLS”, “MYHS”, “MYLS”, “LYHS” and “LYLS” indicate “high yield and high stable”, “high yield and low stable”, “medium yield and high stable”, “medium yield and low stable”, “low yield and high stable” and “low yield and low stable”, respectively. High, medium and low yield is with reference to the center of “mean vs. stability” view of GGE biplot (0,0), which represents the mean yield.
could be due to positive effects of ZT on soils specially when the organic matter content of the soils of the study sites is low and is one of the major reasons for deteriorating soil health in this region [67] [68] . In CT, intensive and shallow tillage leads to significant negative impact on soil health and quality parameter, whereas ZT facilitate buildup in soil organic matter, improve soil biological activity and soil structure due to maintenance of soil aggregates, and reduced oxidation of soil organic matter. These findings are in agreement with [69] and [70] . Results of the long-term research by [19] suggests that ZT significantly reduced bulk density and penetration resistance; increased organic carbon, saturated hydraulic conductivity, water stable aggregate, microbial biomass carbon and soil enzyme activities compared to CT. Additionally, ZT facilitates advance planting (ZT drills works well on relatively high soil moisture which is not the case for CT), regulates (buffers) soil as well as canopy temperature that helps in developing better root system (mass, density, length) which in turn results into better nutrient uptake and plant withstanding against high velocity winds at grain filling/maturity (reduce lodging) [71] .
Based on the mean predictive yield, the six genotypes tested across locations were grouped into high, medium, and low predictive yielders. In both management practices (CT and ZT), high, medium and low predictive yielding genotypes were “HD-2967” and “Munal”; “BAZ”, “CSW-16” and “DPW-621-50”; and “CSW-18”, respectively. In CT, high and medium predictive yielding genotypes were grouped into two separate divisive clusters, which confirm the differential performance of genotypes across locations (Figure 4: Panel A). Conversely, in ZT, all high and medium predictive yielding genotypes grouped into 1 cluster (Figure 4: Panel B). The different clustering pattern in CT and ZT suggests the existence of interaction of soil health dynamics due to CT with different genetic composition, plant architecture, and stress tolerant and disease resistance levels of high and medium predictive yielding genotypes (Supplemental Table S1). In ZT, the low predictive yielding genotype “CSW-18” was highly dissimilar among all the tested genotypes and, thus, formed a unique cluster (Figure 4: Panel B). In contrast, the performance profile of “CSW-18” across locations in CT was on par and shared the same cluster (cluster 1) with the medium and high predictive yielding genotypes (“DPW-621-50” and “Munal”, respectively) (Figure 4: Panel A). Genotypes grouped into the same cluster with the duplicate 1 − R2 ratio value provided the same information (redundancy exist) and can be used interchangeably to reduce testing costs and improve efficiency of breeding programs. However, in both CT and ZT, distinct values of 1 − R2 ratio in each cluster indicated the existence of some extent of dissimilarity among genotypes within the cluster (Figure 4: Panel A and Panel B). This finding can be corroborated with the unique genetic makeup of or distinct parents being used in each genotype (Supplemental Table S1). A definitive conclusion on the most representative and distinct genotype within the cluster must be based on the low value of 1 − R2 ratio. Thus, “Munal” and “HD-2967” were the most representative and distinct genotype in cluster 1 and cluster 2, respectively, in CT (Figure 4: Panel A). Similarly, “HD-2967” was the most representative and distinct genotype in ZT (Figure 4: Panel A). In CT and ZT, the most representative and distinct genotype(s) had the highest predictive yield (Figure 4: Panel A and Panel B). These findings suggest that genetic composition of “HD-2967” has the ability to outperform regardless of alteration made in soil health due to tillage practice. Therefore, growers must select these high performing genotypes as their preferred varieties according to tillage practice and cropping system is followed in their region. Similarly, wheat breeders can use them as a diverse parent for future breeding purposes while considering tillage practice into account so that actual effect of yield can be exploited.
Based on multiple stability measures, wheat genotypes were classified into three categories. Category 1 genotypes had high predictive yield and high stability, and are widely adopted across varied environmental conditions. These genotypes had bi equal to unity, low or non-nonsignificant ![]() and
and![]() , YSi higher than mean YSi, and close to ideal genotype and near zero projection onto the AEC (vertical axis) on “mean vs. stability” view of GGE biplot. Genotypes “HD-2967” (G14) and “Munal” (G19) qualified in category 1 for management practices CT and ZT, respectively (Figure 4: Panel A and Panel B). Significant
, YSi higher than mean YSi, and close to ideal genotype and near zero projection onto the AEC (vertical axis) on “mean vs. stability” view of GGE biplot. Genotypes “HD-2967” (G14) and “Munal” (G19) qualified in category 1 for management practices CT and ZT, respectively (Figure 4: Panel A and Panel B). Significant ![]() of both genotypes contrasts to the previous study where non-significant
of both genotypes contrasts to the previous study where non-significant ![]() was detected for category 1 genotypes [6] . This contrast could be because the errors associated with the slopes of genotypes are not statistically independent [72] . Category 2 genotypes exhibited high predictive yield but low stability. These genotypes had bi greater than unity, high or non-nonsignificant
was detected for category 1 genotypes [6] . This contrast could be because the errors associated with the slopes of genotypes are not statistically independent [72] . Category 2 genotypes exhibited high predictive yield but low stability. These genotypes had bi greater than unity, high or non-nonsignificant ![]() and
and![]() , and high projections onto the AEC (vertical axis) on “mean vs. stability” view of GGE biplot. Thus, these genotypes are suited for specific locations. Results suggest that genotypes “HD-2967” (G14) and “Munal” (G19) were sensitive to environmental change and had greater specificity of adaptability to high yielding environment under management practices ZT and CT, respectively (Figure 1: Panel C and Panel D). Plant breeders can use these genotypes to develop high performers for specific location and management practices. However, according to YSi, category 2 genotypes were better than average and should be considered as stable. Unlike category 1 genotypes, projection onto AEC on “mean vs. stability” view of GGE biplot, category 2 genotypes had relatively opposite projections. This finding confirms the differential performance of the same genotypes in different management practices. Change of stability (high to low or vice versa) of “HD-2967” (G14) and “Munal” (G19) as tillage practices flipped (CT to ZT or vice versa) suggested that the expression of genes governing yield were dependent on management mediated soil health and related processes. Yield of “HD-2967” (G14) was less influenced when soil had higher bulk density, poor water retention capacity, higher hydraulic conductivity, lower soil organic carbon and lower biological activities (soil properties found under CT, [39] . Thus, growers can use “HD-2967” for consistent performance across locations and in cropping system where CT is practiced. In contrast, “Munal” (G19) is recommended for high yield and high adaptability at specific location where CT is practiced or soil had higher bulk density, poor water retention capacity, higher hydraulic conductivity, lower soil organic carbon and lower biological activities. However, growers have to be careful in using location specific genotypes as they had greater specificity of adaptability to high yielding environments. Meaning small change in favorable environment (CT type soils) will result into large fluctuation in genotype performance. Similarly, genotype “Munal” (G19) and “HD-2967” can be used for consistent performance across and within a specific location(s), respectively, and in cropping system where ZT is practiced. Likewise, breeders have scope to further exploit the genes responsible for differential performance of these high yielding and high stable genotype as soil physical, chemical and biological properties are changed due to optimization of tillage practice. Category 3 genotypes had low to medium predictive yield and low stability. These genotypes are suitable for traits (other than yield) such as disease or lodging resistance, where low values are desired in high yielding environments. Category 3 genotypes include “CAW-16” and “CSW-18” in both management practices.
, and high projections onto the AEC (vertical axis) on “mean vs. stability” view of GGE biplot. Thus, these genotypes are suited for specific locations. Results suggest that genotypes “HD-2967” (G14) and “Munal” (G19) were sensitive to environmental change and had greater specificity of adaptability to high yielding environment under management practices ZT and CT, respectively (Figure 1: Panel C and Panel D). Plant breeders can use these genotypes to develop high performers for specific location and management practices. However, according to YSi, category 2 genotypes were better than average and should be considered as stable. Unlike category 1 genotypes, projection onto AEC on “mean vs. stability” view of GGE biplot, category 2 genotypes had relatively opposite projections. This finding confirms the differential performance of the same genotypes in different management practices. Change of stability (high to low or vice versa) of “HD-2967” (G14) and “Munal” (G19) as tillage practices flipped (CT to ZT or vice versa) suggested that the expression of genes governing yield were dependent on management mediated soil health and related processes. Yield of “HD-2967” (G14) was less influenced when soil had higher bulk density, poor water retention capacity, higher hydraulic conductivity, lower soil organic carbon and lower biological activities (soil properties found under CT, [39] . Thus, growers can use “HD-2967” for consistent performance across locations and in cropping system where CT is practiced. In contrast, “Munal” (G19) is recommended for high yield and high adaptability at specific location where CT is practiced or soil had higher bulk density, poor water retention capacity, higher hydraulic conductivity, lower soil organic carbon and lower biological activities. However, growers have to be careful in using location specific genotypes as they had greater specificity of adaptability to high yielding environments. Meaning small change in favorable environment (CT type soils) will result into large fluctuation in genotype performance. Similarly, genotype “Munal” (G19) and “HD-2967” can be used for consistent performance across and within a specific location(s), respectively, and in cropping system where ZT is practiced. Likewise, breeders have scope to further exploit the genes responsible for differential performance of these high yielding and high stable genotype as soil physical, chemical and biological properties are changed due to optimization of tillage practice. Category 3 genotypes had low to medium predictive yield and low stability. These genotypes are suitable for traits (other than yield) such as disease or lodging resistance, where low values are desired in high yielding environments. Category 3 genotypes include “CAW-16” and “CSW-18” in both management practices.
In BR, high predictive yielding genotypes tended to perform better in ZT over CT, whereas low predictive yielding genotypes performed better in CT. In both CT and ZT, different genotypes constituted the three clusters, which suggest the differential performance of genotypes across years and management practices within a location. In CT, genotypes “CSW-16”, “HD-2824”, “K-307”, “DPW- 621-50” and “HD-2967” had high predictive yield and stability across years based on multiple stability measures. These genotypes had greater adaptability in this location under management practice CT. Conversely, genotypes “DPW-621- 50”, “HD-2733”, and “Munal” had high predictive yield and stability in ZT. Wheat researchers can use high predictive yielding, high stable and low 1 − R2 ratio genotypes from different clusters as a contrast parent for future breeding purposes. These genotypes include “HD-2824” and “DPW-621-50”, and “Munal” in CT and ZT, respectively.
Similar to BR, in MP, high and low predictive yielding genotypes tended to perform better in ZT and CT, respectively. Relatively, CT resulted in more diverse performances of ten tested genotypes, thus forming 5 clusters. Genotypes “HD-2967”, “Munal”, “BAZ” and “MPO-1215” had medium to high predictive yield and stable across years in CT. The significant bi value of “Munal” suggested that its performance improved in good years. In ZT, medium predictive yielding genotypes were stable. These genotypes include “BAZ” and “DPW-621-50”. High predictive yielding genotypes had large prediction intervals and non-non- significant![]() . In CT, genotypes “HD-2932”, “BAZ” and “JW-3288” with the lowest 1 − R2 ratio had high stability. In contrast, in ZT, “GW-322” and “HD- 2967” had the lowest 1 − R2 ratio but with low stability. Pedigree of “GW-322” confirmed the presence of stem rust (Sr2, Sr11+), leaf rust (Lr1, Lr10, Lr13, Lr13+), yellow rust (Yr2+) and glutenin (Glu-A1b, Glu-B1b, Glu-D1a) genes (Supplemental Table S1). Glutenin (a type of glutelin) is the major protein of wheat flour. Similarly, “BAZ” and “HD-2932” have adult plant stem rust (Sr2+) and leaf rust (Lr13+) resistant genes, respectively. Additionally, previous studies have identified that “BAZ” and “GW-322” have capability to withstand heat and drought stress. Therefore, researchers can introgress genes from these genotypes into elite inbreds to make better hybrids with high stability. In PB, a small percentage of the total variation from GxM and GxMxY interactions (p < 0.01) suggests that the grain yield variation from these interactions is the least important. Furthermore, estimates of random effects of genotypes revealed mixed response of high and low predictive yielding genotypes in both management practices. Similarly, a divisive cluster did not identify dissimilarity among genotypes, except for genotype “DBW-88” in CT. Likewise, genotypes “DPW-621-50”, “HD- 2967” and “PBW-550” had high predictive yield and high stability in both CT and ZT. The mixed response of high and low predictive yielding genotype could be due to dominant nature of complex interaction of low solar radiation and low temperature with pre-anthesis over plant growth-tillage practice interaction. The combined effect of temperature and radiation influence plant process differently. High temperature and low radiation during the phase of rapid spike growth and development (20 - 30 days prior anthesis) result in reduced number of grains m−2 and thousand kernel weight [73] . There is a general consensus among the scientific community that the global temperature is rising due to climate change, and the largest increase in mean temperature is projected for the high latitude regions [74] . PB is relatively situated at higher latitudes and, thus, it might have experienced more temperature fluctuations. Temperature and solar radiation variation among locations tested are confirmed in Supplemental Figure S1.
. In CT, genotypes “HD-2932”, “BAZ” and “JW-3288” with the lowest 1 − R2 ratio had high stability. In contrast, in ZT, “GW-322” and “HD- 2967” had the lowest 1 − R2 ratio but with low stability. Pedigree of “GW-322” confirmed the presence of stem rust (Sr2, Sr11+), leaf rust (Lr1, Lr10, Lr13, Lr13+), yellow rust (Yr2+) and glutenin (Glu-A1b, Glu-B1b, Glu-D1a) genes (Supplemental Table S1). Glutenin (a type of glutelin) is the major protein of wheat flour. Similarly, “BAZ” and “HD-2932” have adult plant stem rust (Sr2+) and leaf rust (Lr13+) resistant genes, respectively. Additionally, previous studies have identified that “BAZ” and “GW-322” have capability to withstand heat and drought stress. Therefore, researchers can introgress genes from these genotypes into elite inbreds to make better hybrids with high stability. In PB, a small percentage of the total variation from GxM and GxMxY interactions (p < 0.01) suggests that the grain yield variation from these interactions is the least important. Furthermore, estimates of random effects of genotypes revealed mixed response of high and low predictive yielding genotypes in both management practices. Similarly, a divisive cluster did not identify dissimilarity among genotypes, except for genotype “DBW-88” in CT. Likewise, genotypes “DPW-621-50”, “HD- 2967” and “PBW-550” had high predictive yield and high stability in both CT and ZT. The mixed response of high and low predictive yielding genotype could be due to dominant nature of complex interaction of low solar radiation and low temperature with pre-anthesis over plant growth-tillage practice interaction. The combined effect of temperature and radiation influence plant process differently. High temperature and low radiation during the phase of rapid spike growth and development (20 - 30 days prior anthesis) result in reduced number of grains m−2 and thousand kernel weight [73] . There is a general consensus among the scientific community that the global temperature is rising due to climate change, and the largest increase in mean temperature is projected for the high latitude regions [74] . PB is relatively situated at higher latitudes and, thus, it might have experienced more temperature fluctuations. Temperature and solar radiation variation among locations tested are confirmed in Supplemental Figure S1.
For the development of stable genotype for grain yield, the pattern of GxExM interaction, grouping of genotypes into divisive clusters and estimates (with prediction intervals) of genotype varied in management practice CT and ZT. Based on the results, it is possible to breed tillage management specific stable genotypes with enhanced stress and quality traits. In dark zones of Indo-Gangetic plains (areas where water table is declining due to overexploitation of ground water resources), the Government of India has been emphasizing on diversification of rice with maize and CT with ZT in rice-wheat cropping system [61] and [19] . Thus, growers can chose tillage practice specific high yielding and high stable genotypes in their cropping system. Across locations, the genotypes “Munal” and “HD-2967” were the best performers and high stable in CT and ZT, respectively. Genotypes “HD-2824” and “DPW-621-50”, and “Munal” may serve as diverse parents for developing high quality climate smart, locally adapted genotypes for BR in CT and ZT, respectively. Similarly, genotypes “HD-2932”, “BAZ” and “JW-3288”, and “GW-322” and “HD-2967” can be used for research purposes for developing locally adapted, stress tolerant genotypes for MP in management practices CT and ZT, respectively.
Supplemental Material Available
Supplemental material is available with the online version of this article.
Acknowledgements
This research was funded by Die Bundesministerium für Wirtschaftliche Zusammenarbeit und Entwicklung (BMZ/GIZ), Germany. The land and other facilitites for field trials, provided by Borlaug Institute for South Asia (BISA), are thankfully acknowledged. We would like to thank CGIAR Research Program on Wheat Agri-Food Systems (WHEAT) for supporting this study. We express our appreciation to Dr Hans-Joachim Braun, Dr. Ravi P. Singh and Dr. Arun K Joshi of CIMMYT, Mexico, for their support in this research. We would also like to thank Dr. Mahendra Dia from North Carolina State University, NC, USA, for providing his subject matter expertise and support in statistical analysis using SAS and R programming language.
Abbreviations
AEC = Average environment coordinate
ANOVA = Analysis of variance
BLUP = Best linear unbiased predictor
BR = Pusa, Bihar
GGE = Genotype main effects plus genotypic × environment interaction effect
GGL = Genotype main effects plus genotypic x location interaction effect
GxE = Genotype x environment interaction
GxMxE or GxMxLxY = Genotype x management × environment interaction or genotype x management
x location x year interaction
G = Genotype
G01 or 1 = BAZ
G02 or 2 = CSW-16
G03 or 3 = CSW-18
G04 or 4 = DBW-17
G05 or 5 = DBW-88
G06 or 6 = DPW-621-50
G07 or 7 = GW-273
G08 or 8 = GW-366
G09 or 9 = GW-322
G10 or 10 = HD-2824
G11 or 11 = HD-2687
G12 or 12 = HD-2733
G13 or 13 = HD-2932
G14 or 14 = HD-2967
G15 or 15 = HI-1563
G16 or 16 = JW 3288
G17 or 17 = K-307
G18 or 18 = MPO-1215
G19 or 19 = Munal
G20 or 20 = PBW-343
G21 or 21 = PBW-550
HYHS = High yield and high stable
HYLS = High yield and low stable
L = Location
LYHS = Low yield and high stable
LYLS = Low yield and low stable
M = Management
MET = Multi-environment trial
MP = Jabalpur, Madhya Pradesh
MYHS = Medium yield and high stable
MYLS = Medium yield and low stable
PB = Ludhiana, Punjab
PC = Principal component
RGxE = R language program for the analysis of genotype stability and location value
REML = Restricted maximum likelihood
SASGxE = SAS program for the analysis of genotype stability and location value
SVP = Singular value partitioning
Y = Year
![]() or Si = Shukla’s stability variance
or Si = Shukla’s stability variance
bi = Linear regression coefficient
![]() = Deviation from regression
= Deviation from regression
YSi = Kang’s stability statistic
Supplemental Material Description
The Supplemental tables provide pedigree, phenotypic and genotypic characteristics, and univariate statistics of tested genotypes for 3 years at 3 locations.
The supplemental figures provide meteorological data of location BR, MP and PB for the year 2012 to 2015, yearly BLUP-genotype main effects plus genotype x location interaction effect (BLUP-GGL) biplot and mean (95% CI) for wheat yield (t ha−1) of 6 genotypes tested in 3 years and 3 locations over 2 management practices.
Supplementary Table 1. The 21 wheat genotypes tested with pedigree, phenotype and genotypic information.
Source: Wheat Atlas―http://www.wheatatlas.org/ # “A” = tested in all location for 3 yrs; “B” = tested only in Bihar (BR) for 3 yrs; “M” = tested only in Madhya Pradesh (MP) for 3 yrs; “P” = tested only in Punjab (PB) for 3 yrs; “X” = tested only in MP for 1 yrs; “Y” = tested only in PB for 1 yrs. †Under participatory trials and demonstration since 2011. ‡Registered with PPVFRA (Protection of Plant Varieties and Farmers Right Authority, India). § “*” = backcross, “/” = single cross, “//” = double cross. ¶ Maturity: Early (E), <120 days; medium (M), 120-140 days; late (L), >140 days. ‡‡ Height: Short (S), <90 cm; medium (M), 90-100 cm; Tall (T), >100 cm. †† Yield potential. §§ 1000 kernel weight. ¶¶ Grain characteristics: White and bold (WB); bold, medium hard and lustrous (BMHL); long and hard (LH); amber, medium bold, hard and lustrous (AMBHL); amber, hard and medium size (AHM), amber semi-hard and bold (ASB); amber hard and bold (AHB). ## Wheat type: Bread wheat (B); Durum wheat (D). ††† “Y” = Yes; “N” = No; “-” = No info; “H” = heat; “VD” = variable sowing dates; “D” = drought; “L” = lodging.
Supplementary Table 2. Significance value of regression coefficient, deviation from regression, Shukla’s stability variance, and Kang’s stability statistics for wheat yield (t∙ha−1) of 6 genotypes tested in 3 years and 3 locations over 2 management practices; and for location BR, MP, and PB for wheat yield (t∙ha−1) of 10 (6 same + 4 different) wheat genotypes tested for 3 years over 2 management practices.
*, ** and *** significantly different from unity for the regression coefficients or slope (bi) and from zero for the deviation from regression (S2d) and Shukla’s stability variance (σi2) at 0.05, 0.01 and 0.001 levels of probability, respectively. “§” indicates negative slope. “√” indicates stable according to Kang stability statistics (YSi).
![]()
Supplementary Figure 1. Daily average temperature (˚C), Solar Radiation (MJ∙m−2∙day−1) and average precipitation (mm∙day−1) for location BR, MP and PB for the year 2012 to 2015. Vertical dashed line represents date of sowing (Nov 15). Vertical solid red and blue lines represent harvest date for location BR and MP, and PB, respectively. (Source: NASA, USA).
![]()
Supplementary Figure 2. Daily wind speed (m∙s−1), relative humidity (%) and dew/frost point temperature (˚C) for location BR, MP and PB for the year 2012 to 2015. Vertical dashed line represents date of sowing (Nov 15). Vertical solid red and blue lines represent harvest date for location BR and MP, and PB, respectively. (Source: NASA, USA).
![]()
Supplemental Figure 3. The polygon (which-won-where) view of BLUP-genotype main effects plus genotype × location interaction effect (BLUP-GGL) biplot for year 2012 (Panel A and Panel B), 2013 (Panel C and Panel D) and 2014 (Panel E and Panel F) for yield of 6 genotypes tested in 3 years and 3 locations over 2 management practices. The biplots were based on Scaling = 0, Centering = 0, and SVP = 2. Key to the labels of genotype and location is presented in abbreviation section.
![]()
Supplemental Figure 4. The mean vs. stability view of BLUP-genotype main effects plus genotype x location interaction effect (BLUP-GGL) biplot for year 2012 (Panel A and Panel B), 2013 (Panel C and Panel D) and 2014 (Panel E and Panel F) for yield of 6 genotypes tested in 3 years and 3 locations over 2 management practices. The biplots were based on Scaling = 0, Centering = 0, and SVP = 2. Key to the labels of genotype and location is presented in abbreviation section.
![]()
Supplemental Figure 5. Mean (95% confidence interval) yield of 6 genotypes tested in 9 environments (3 locations × 3 years) over 2 management practices (CT and ZT). The shaded band represents 95% confidence interval. The environment 1, 2, 3, 4, 5, 6, 7, 8, and 9 represent BR-2012, BR-2013, BR-2014, MP-2012, MP-2013, MP-2014, PB-2012, PB-2013, and PB-2014, respectively.
![]()
Submit or recommend next manuscript to SCIRP and we will provide best service for you:
Accepting pre-submission inquiries through Email, Facebook, LinkedIn, Twitter, etc.
A wide selection of journals (inclusive of 9 subjects, more than 200 journals)
Providing 24-hour high-quality service
User-friendly online submission system
Fair and swift peer-review system
Efficient typesetting and proofreading procedure
Display of the result of downloads and visits, as well as the number of cited articles
Maximum dissemination of your research work
Submit your manuscript at: http://papersubmission.scirp.org/
Or contact ajps@scirp.org