1. Introduction
Over two decades ago, we started research investigating air-pollutant-philic plants that can grow with air pollutants as the sole nutrient source [1,2]. We thought (and still think) that such plants are critical for mitigation of air pollution. Our idea is a critical response to the need to cope with harsh air pollution worldwide and is a viable possibility due to plants’ natural ability as a sink of various air pollutants. Although we still have much ground to cover and are not as far along as we had had hoped, we have made some progress in the research of air-pollutantphilic plants.
We have primarily focused on atmospheric nitrogen dioxide (NO2) as a target pollutant. The weighted average of NO2 concentration in cities from over 141 countries is reported to be 50.6 µg/m3 (≈27 ppb) [3]. The current WHO air quality guideline value (annual mean) for NO2 is 40 µg/m3 (≈21 ppb) [4].
The air in polluted urban areas of Japan contains about 0.05 ppm of NO2, a similar or even higher amount of nitrogen oxide (NO), and 0.1 ppm of non-methane hydrocarbons (NMHCs) [5]. Methane and other volatile organic hydrocarbons, including NMHCs, photochemically produce hydroxyl radicals (highly reactive molecules containing a single unpaired electron), which react with nitrogen oxides (NOx) to produce photooxidative ozone. NO2 and oxidants remain in the lower atmosphere (troposphere) and are very toxic gases for animals, including humans, as well as for plants [6]. Gaseous pollutants such as NOx, once emitted, disperse into the atmosphere rapidly. Centralized treatment of these scattered pollutants is energy (equivalent to the entropy) intensive and costly. In contrast, plants are driven by solar energy, are self-reproducing, and can concentrate and detoxify pollutants. Therefore, plants should prove to be very useful in helping to clean up dispersed, ambient pollutants. In fact, we know that plants do uptake NO2 [7-11] and incorporate its nitrogen into organic nitrogenous compounds [12-15].
2. Variation in Plants’ Abilities to Mitigate Air Pollutants among Naturally Occurring Plants
Learning to what extent naturally occurring plants are (or are not) air-pollutant-philic is important. We therefore investigated the uptake and assimilation of NO2 in 217 taxa of naturally occurring plants that included 107 woody plants, 60 cultivated herbaceous plants, and 50 wild herbaceous plants collected from roadsides.
Plants were fumigated in a fumigation chamber at 22˚C ± 0.3˚C, 70% ± 4% relative humidity, and an atmospheric level of 0.03% to 0.04% CO2 for 8 h during daytime (09:00 - 17:00) under fluorescent lights (70 µmol photons m–2·s–1) with 4 ± 0.4 ppm 15NO2 (51.6 atom% 15N). Leaves were then cut from fumigated plants, washed with tap water and then with distilled water, and lyophilized. Lyophilized leaves were then analyzed for the 14N plus 15N content and the percentage of 15N in the samples was determined using an EA-MS analyzer in which an elemental analyzer (EA/NA; Fisons Instruments, Milano, Italy) interfaced with a mass spectrometer (Delta C; Thermo Finnigan, Bremen, Germany), as described previously [16]. From these two values, total (14N plus 15N) nitrogen derived from NO2 in the leaves of fumigated plants (mg N/g dry weight) was determined (designated TNNO2). Parts of the lyophilized leaves were subjected to Kjeldahl digestion [17,18], and the reduced or Kjeldahl nitrogen fraction was obtained. The 14N plus 15N content and the percentage of 15N in the Kjeldahl fraction were then analyzed. From these values, reduced nitrogen derived from NO2 in the leaves of fumigated plants (mg N/g dry weight) was determined (designated RNNO2). TNNO2 (mg N/g dry wt), RNNO2 (mg N/g dry weight), and the ratio of RNNO2/RN (%) respectively give a measure of the ability of plants to act as a sink for NO2, that of the ability of plants to metabolize (assimilate) NO2, and of the contribution of NO2 as a nitrogen source.
Details of the data on 217 taxa have been published [16,19], and here we briefly discuss some points in relation to the air-pollutant-philic plants. Among the 217 taxa, the RNNO2 differed by a factor of 657 between the highest (Eucalyptus viminalis, 6.57) and lowest (Tillandsia ionantha and Tillandsia caput-medusae, 0.01). These two species belong to the Myrtaceae and Bromeliaceae family, respectively. The Myrtaceae seems to include plants with a high ability to assimilate NO2 (see below), and plants of the Bromeliaceae have less of an ability to assimilate NO2. The other two species of the Bromeliaceae also showed low values for RNNO2: 0.02 and 0.12 in Tillandsia usneoides and Tillandsia geminiflora, respectively.
The RNNO2/RN differed about 85-fold between the highest (Magnolia kobus, 12.7%) and lowest (Codiaeum variegatum, 0.15%). These two species belong to the Magnoliaceae and Euphorbiaceae family, respectively. The 217 taxa examined included no other species of the Magnoliaceae. One species of the 217 taxa, belonging to the Euphorbiaceae, Sapium sebiferum, showed a particularly high value of RNNO2/RN (10.1) [16].
Among the 217 taxa, six woody species, one cultivated herbaceous species, and two wild herbaceous species collected from roadsides were found to have RNNO2 greater than 4 and RNNO2/RN values greater than or close to 9 as shown in Table 1. These nine species may have more potential to be air-pollutant-philic specimens than those examined in the 217 taxa examined to date.
Since the fumigation in our study described above lasted for 8 h, RNNO2/RN values reflect the turnover rate of reduced nitrogen in plants. Turnover of reduced nitrogen is considered to be first order as in the case of proteins [20]. A simple exponential curve was fitted to the RNNO2/RN data to generate a first-order rated constant: k. The half-life of the reduced nitrogen was calculated as
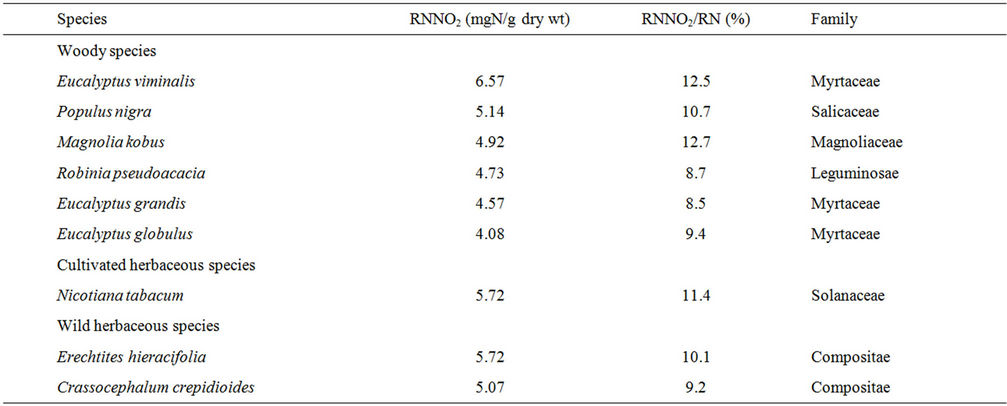
Table 1. Nine species with high values of reduced nitrogen derived from NO2 (RNNO2) and the corresponding percentage of RNNO2 in the total reduced nitrogen (RNNO2/RN).
t1/2 = ln(2)/k. In the aforementioned nine species, the half-life of reduced nitrogen for M. kobus with the highest RNNO2/RN value (see Table 1) was calculated to be approximately 40 h. This value is comparable to the half-life of Cullin-associated and neddylation-dissociated 1 (CAND1; 30 h) and that of Transport inhibitor response 1 (TIR1; 10 h) proteins [20]. However, assuming a linear dependence of the turnover rate on the concentration of NO2, the half-life of the reduced nitrogen at 0.04 ppm is estimated to be 4000 h. More recently, we found that 14% of TNNO2 was derived from NO2 in cucumber (Cucumis sativus) plants grown under natural lighting in the presence of 100 ± 50 ppb NO2 for 5 weeks [21]. Using these values and the assumptions described above, the half-life of total nitrogen in cucumber was estimated to be 1000 h. Obviously, these calculations of half-lives of nitrogen-derived from NO2 in the selected species indicates that no naturally occurring plants studied to date are air-pollutant-philic. However, the existence of great differences in both RNNO2 and RNNO2/RN is encouraging for the fabrication of air-pollutant-philic plants. Investigations on the causes for the differences in these parameters between species or how the metabolism of NO2 differs between species may provide an important clue to improve plants’ abilities to assimilate NO2 and hence to make plants air-pollutant-philic agents.
In the 217 taxa examined, the RNNO2 value varied among the individuals of a species [16]. This variation was more pronounced in the wild herbaceous and woody plants than in the cultivated herbaceous plants. Among wild herbaceous plants (19 individuals), the variation was 26.2-fold in Solidago altissima, 21.4-fold in Rumex acetosella, and 17.6-fold in Crassocephalum crepidioides. In woody plants, the corresponding value was 11.9-fold in M. kobus, 10.6-fold in Euonymus japonicus, and 6.7-fold in Prunus cerasoides [16]. Understanding differences among individuals of the same species in their abilities to assimilate NO2 is also important in terms of genetically constructing air-pollutant-philic plants.
We attempted selection of “mutant” lines of Rhododendron mucronatum, which is widely used as roadside trees and can be propagated by cuttings [22]. R. mucronatum was ranked 105th among the 217 taxa in RNNO2. In 1996, 1452 different cutting lines of R. mucronatum (referred to as the “96-plants”) were fumigated with 4 ppm 15NO2 for 8 h, and TNNO2 in the leaves was determined. A 56-fold difference was observed between the individuals with the highest and lowest capacities. We propagated approximately 100 lines (designated the “97- plants”) each from the high-, middle-, and low-capacity groups, and assayed them for TNNO2 in the following year. The maximum variation in TNNO2 was 5.6 times. In 1998, 227 lines propagated from the 96-plants and 95 lines from the 97-plants (the “98-plants”) were assayed for TNNO2. The variations in TNNO2 in the 97- and 98- plants were maximally 4.9 and 5.2 times, respectively. The reason(s) for the disappearance of the initial large variation in TNNO2 remains unclear.
3. Genetic Improvement of Plants’ Abilities to Mitigate Air Pollutants
Foliar uptake of NO2 is believed to occur via stomatal and nonstomatal pathways as a gas or as nitrite or nitrate [11]. However, the precise mechanisms remain unclear. Most NO2 molecules taken up into plants are converted, either chemically or enzymatically, to nitrate and nitrite, assimilated to ammonia, and incorporated into amino acids and other organic nitrogenous compounds through the primary nitrate assimilation pathway [12-15]. In the primary nitrate pathway, nitrate is converted to nitrite by a two-electron reduction catalyzed by the nitrate reducetase (NR), the first enzyme. The second enzyme, nitrite reductase (NiR), catalyzes a six-electron reduction to convert nitrite to ammonia. Ammonia thus formed is incorporated into glutamic acid to form glutamine by the glutamine synthetase (GS), the third enzyme. Therefore, engineering of these genes should be instrumental to improve metabolism of NO2 and help make plants air-pollutant-philic.
To date we have produced transgenic plants of Arabidopsis thaliana (Cruciferae), a model species that overexpresses any one of the three genes of the primary nitrate reduction pathway [17]. We have also produced transgenic plants of Rhaphiolepis umbellata (Rosaceae) that overexpress the NiR gene [23]. R. umbellata is a very popular roadside shrub in Japan. Finally, we produced mutants of Ficus pumila (Moraceae) using an ion-beam-irradiation technique [24], an emerging method to produce various mutants [25]. F. pumila is an evergreen climber that is sometimes used to cover the walls of houses and buildings. Data on the uptake and assimilation of NO2 by these three genetically modified plants are shown in Table 2.
The uptake of NO2 in transgenic R. umbellata and ion-beam-irradiated F. pumila was 1.8 to 1.9 times greater compared with their respective control plants (Table 2). Although these increases in the uptake of NO2 may not be enough to make these plants air-pollutantphilic, note that engineering of the NiR gene and ionbeam irradiation in F. pumila did improve the plants’ abilities to uptake NO2. Future studies investigating what causes the increase in uptake of NO2 in these modified plants may provide some information that is helpful in making plants air-pollutant-philic.
The assimilation of NO2 in transgenic A. thaliana and R. umbellata, and ion-beam-irradiated F. pumila, was 1.6 -
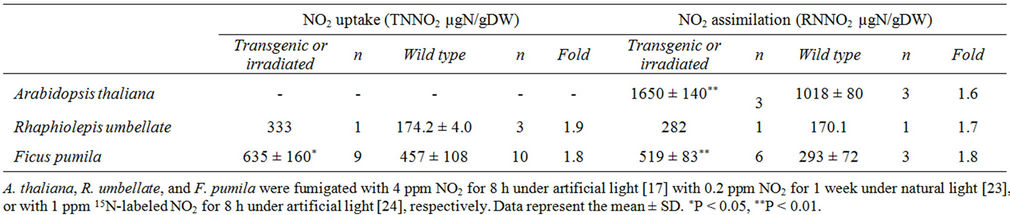
Table 2. Total nitrogen derived from NO2 (TNNO2) and reduced nitrogen derived from NO2 (RNNO2) in transgenic Arabidopsis thaliana and Rhaphiolepis umbellata plants, and plants regenerated from ion-beam-irradiated explants of Ficus pumila.
1.8 times greater compared with their respective control plants (Table 2). Thus, gene engineering of the NiR gene and ion-beam irradiation improved plants’ abilities to assimilate NO2. This finding was in contrast to the effects of overexpressing NR and GS (see below). In transgenic A. thaliana, a 1.5-time increase in NiR enzyme activity was observed as compared to the control [17]. The increase in the ability of assimilating NO2 in transgenic A. thaliana can therefore be attributed to the increase in NiR activity. However, no such increase in NiR activity was observed in ion-beam-irradiated F. pumila [24]. This implies that genetic change(s) other than manipulation of the NiR gene may be capable of improving the assimilation of NO2. Future studies on the molecular causes of improved NO2 assimilation in ion-beam-irradiated F. pumila may provide a clue to help create airpollutant-philic plants.
In the case of the overexpression of NR and GS in A. thaliana, no significant increases in the uptake or assimilation of NO2 was observed despite respective enzyme activities having been increased by 1.8- and 1.5-fold compared to the controls [17]. Taken together with the above results on NiR overexpresssion, NiR, but not NR or GS, is a controlling enzyme of NO2 assimilation in plants. This conclusion was supported by the determination of the flux control coefficient, a measure of the effect of change in a single enzyme activity on flux [26,27] as described previously [17].
4. Plant Vitalization Effect of NO2
During our study on air-pollutant-philic plants, we unexpectedly discovered that prolonged exposure of plants to a sufficient level of NO2 activates the uptake and metabolism of nutrients that fuel plant growth and development [28]. We named this phenomenon the plant vitalization effect of NO2 (PVEON).
In the literature, atmospheric NO2 has been regarded as being either detrimental [15,29] or beneficial [30,31] to plants depending on the plant’s nitrogen nutrient conditions. Recent reports that NO2 stimulates or fails to stimulate the growth of a particular plant [32,33] suggest that this debate is still active. We speculate that a cause for this discrepancy may be that the response of plant growth to NO2 is highly dependent on the target species, and thus both the concentration of NO2 and duration of exposure to NO2 need to be optimized for each plant species to observe the PVEON.
The PVEON was first found in Nicotiana plumbaginifolia [28], and then in other species such as cucumber, kenaf, lettuce, pumpkin, and tomato (Table 3) [21,28,34, 35]. Shoot biomass in NO2-exposed plants (+NO2) was 1.4- to 2.4-fold greater than that in non-exposed (–NO2) plants. A. thaliana showed PVEON and its increase in shoot biomass in response to this effect differed between the accessions Columbia and C24 (1.6- and 2.3-fold, respectively) compared to no-exposure plants when plants were exposed to 50 ppb NO2 for 3 weeks [35]. The optimal NO2 concentration for the PVEON varied from 50 to 200 ppb depending on the species. The duration of exposure to NO2 was changed from 3 to 12 weeks as a function of target species (Table 3). The increase in shoot biomass in response to PVEON was always accompanied by an increase in total leaf area in all plant species studied to date. In Nicotiana plumbaginifolia, the contents per shoot of C, N, S, P, K, Ca, Mg, free amino acids and crude proteins were almost doubled in +NO2 plants compared with –NO2 plants [28].
In tomato (cv. Micro-Tom), the PVEON significantly increases the yield of fruits (Table 3). This increase in fruit yield was accompanied by acceleration of flowering time by 3.2 days and an increase in flower number per plant of up to 60%. Not surprisingly, the average weight of a tomato fruit was not significantly different between +NO2 plants and –NO2 plants, but the number of fruits on a plant was distinctly increased in the former [35]. Thus, the PVEON increases fruit yield via stimulation of flowering in tomato. This is in sharp contrast to Spierings’ [36] report that exposure of Moneymaker tomato plants to NO2 at 250 ppb during the entire growth period of about 4 months caused a 22% decrease in fruit yield. In addition, Spierings [36] found that NO2 was detrimental to the vegetative growth of Moneymaker; fumigation with NO2 increased the height of very young plants, but resulted in smaller leaves and shorter petioles, and
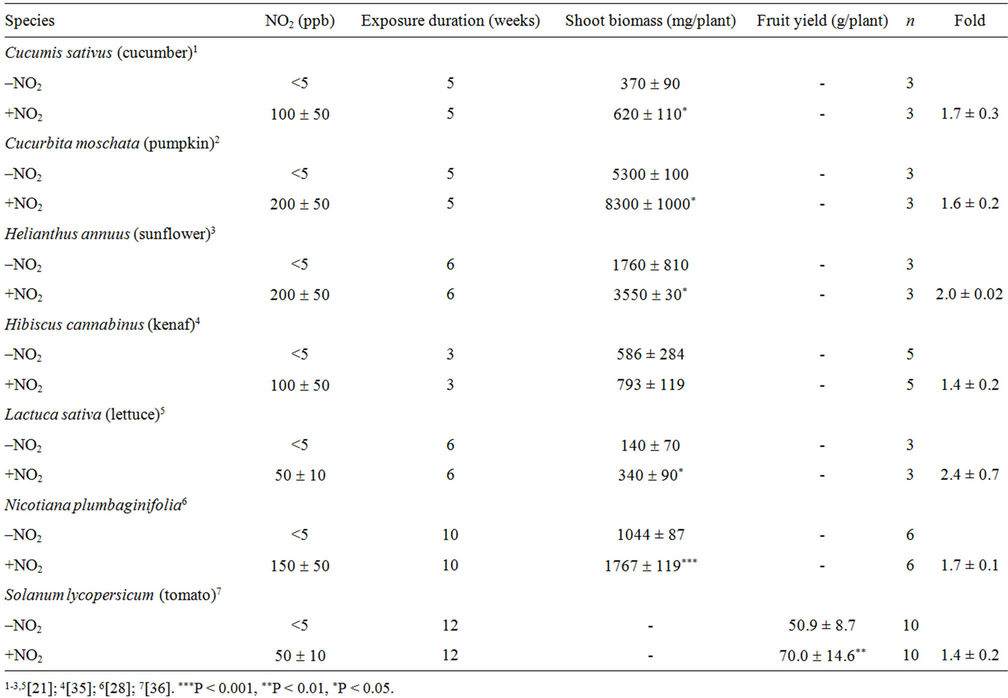
Table 3. Effects of exposure to NO2 on the yield of shoot biomass or fruit.
caused a loss of leaves. These findings are also contrary to our results for Micro-Tom, where no detrimental effect of NO2 on vegetative growth was observed, and the biomass yield of Micro-Tom plants grown with or without 50 ppb NO2 was not significantly different. The reason for these discrepancies remains unclear.
Based on the mass spectrometric analysis on the 15N/14N ratio, it was found that NO2-derived N (NO2-N) comprised less than 3% - 14% of the total plant nitrogen [21,28,34], indicating that PVEON is due to the effect of NO2 as a signal rather than as a nitrogen source. This speculation is in line with the common understanding that NO2 is a reactive nitrogen species acting as a cellular signal in both plants and animals.
The molecular mechanisms of PVEON and the genes involved in PVEON are currently being investigated using Arabidopsis plants. Histological analyses examining the causes of an approximately two times increase in leaf size in Arabidopsis plants indicates that an increase in cell size rather than cell number is the most probable cause. Based on transcriptome analysis, we found that destruction of a gene (Vita 1) causes loss of PVEON. Thus, this gene is a PVEON-responsive gene in Arabidopsis plants. These genes will prove useful in helping to make plants air-pollutant-philic in future studies.
5. Acknowledgements
This work was supported in part by a Grant-in-Aid for Scientific Research (nos.18770037 and 21580403) from the Japan Society for the Promotion of Science and by the Research for Promoting Technological Seeds Program of the Japan Science and Technology Agency.