An in vitro infection model system to study proteins expressed during interaction of mycobacterium with murine macrophages ()
1. INTRODUCTION
Tuberculosis (TB), caused by Mycobacterium tuberculosis is a devastating health problem. World Health Organization (WHO) estimates that 2 billion people have latent TB while another 3 million people worldwide die of TB each year [1]. Resurgence of tuberculosis has intensified the necessity for new approaches to combat M. tuberculosis. M. tuberculosis is a facultative, intracellular pathogen which resides within the macrophages of the host. To survive intracellularly it must respond to the intracellular milieu of the macrophages. It has been well documented that intraphagosomal survival and growth of mycobacteria in macrophage is associated with changes in their gene expression and protein composition upon engulfment [2-4].
To date, in spite of advancement in mycobacterial research our knowledge about genes and their corresponding functional proteins involved during the interaction of mycobacterium with macrophages remains fragmentary. Though several research groups have done proteomic studies on M. tuberculosis and other members of the genus [5-7], only few researchers have utilized proteomics to determine changes in protein composition in response to the intraphagosomal environment [3-8]. Such studies have mostly been carried out using cell lines, which definitely have limitations to model in vivo situation. Several reports have demonstrated that the use of different cell lines and culture conditions can significantly influence the protein patterns and intracellular survival of mycobacteria [9,10].
In the present study we have worked to develop an in vitro model using murine peritoneal macrophages and M. bovis BCG. BCG, though an attenuated vaccine strain, retains its ability to survive within the macrophages hence expected to mimic some of its behavior with the pathogenic M. tuberculosis inside the macrophages. Multiplication of the mycobacteria inside the macrophages was ascertained using thymidine uptake assay. SDS-PAGE was employed to study the proteins expressed and selected proteins were subjected to mass spectrometry.
2. MATERIALS AND METHODS
Mycobacteria and culture:
M. bovis BCG (Danish) procured from Mycobacterial Repository Centre at National JALMA Institute for Leprosy and other Mycobacterial Diseases, Agra was cultured in Sauton’s liquid medium at 37˚C. Cells were harvested in late log phase (4 weeks) of growth.
Animals:
BALB/c mice (6-8 weeks old) of 20 ± 2 g of weight were obtained from Institute’s Animal House. The animals were given a standard pellet diet and water ad libitium. The animal studies were strictly performed following mandates approved by the Animal Ethical Committee of the Institute.
Macrophage culture and infection with M. bovis BCG:
BALB/c mice were sacrificed by cervical dislocation and murine peritoneal macrophages were collected as lavage in RPMI 1640 (Sigma). After washing, cells were adjusted to a density of 5 × 104 cells/well (medium with 10% fetal calf serum, 2mM glutamine, 100 µg/ml antibiotic) of a 96-well culture plate which was kept for 48 hrs in a CO2 incubator. Prior to infection with mycobacteria, non-adherent cells were removed and macrophages were suspended in antibiotic free medium. M. bovis BCG cells, sonicated for 15sec to disperse clumps were added to the adhered macrophage monolayer at different infection ratios and left to phagocytose for 12 hrs. After 12 hrs extracellular mycobacteria were removed by extensive washing and the infected macrophages were replenished with medium containing gentamicin (10 µg/ml) to prevent extracellular replication of mycobacteria. Macrophage viability (never fell below 80%) was assessed by trypan blue exclusion. Incubations were carried out for 5 days in CO2 incubator and 18 hrs prior to termination the cultures were pulsed with 1µCi [3H] thymidine (methyl-T, specific activity approx. 17.2 Ci/ mmol from BRIT, India) per well. Cells were harvested on a cell harvester and the radioactivity was measured in a liquid scintillation counter.
Mycobacterial cell lysate proteins:
Cell lysates were prepared according to the recommended protocol [11]. Cells were pelleted by centrifugation, washed and suspended (0.2 g/ml) in sonication buffer (50 mM Tris/HCl, pH 7.4 with 10 mM MgCl2, 1 mM PMSF and 1mM EGTA) and sonicated for 10 min. Lysates were clarified by centrifugation and protein concentration was estimated using Bradford method [12]. For infected cell lysate, macrophages grown in 25 cm2 culture flasks at density of 2-5 × 106 cells/ml were infected with M. bovis BCG at ratio of 1:10. Finally cells were washed, collected in PBS and lysate prepared. Using similar protocol, non infected macrophage cell lysate was also prepared.
Sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE):
Mycobacteria, macrophage and infected cell lysates were subjected to SDS-PAGE. SDS-PAGE under reducing condition was done by the method of Laemmli [13] using 12% resolving gel. An exact amount (25 µg) of protein was loaded in each lane and experiment was repeated atleast three times. After electrophoresis the gels were stained for proteins with silver stain using Investigator Silver Stain Kit (Genomic Solutions).
In-gel digestion with trypsin and mass spectrometry (MS):
Method of Shevchenko et al was followed for in-gel digestion of proteins [14]. Protein spots of interest were excised from gel using Investigator ProPic (Genomic Solutions). Digestion of proteins and spotting of peptides on matrix assisted LASER desorption/ionization-time of flight (MALDI-TOF) target plate was carried out using Investigator ProPrep (Genomic Solutions). Gel plugs destained with K3[Fe(CN)6] and Na2S2O3 solution were treated with 10 mM DTT followed with 55 mM iodoacetamide and incubated for 12 h at 37°C with trypsin (Promega). Samples were then desalted with C-18 ZipTips, which were eluted on target plate with 2 μl of α- cyano-4-hydroxycinnamic acid. Mass spectra were acquired with Autoflex II TOF/TOF 50 (Bruker Daltonics) in positive reflectron mode, in the detection range of 500-3000 m/z. Peak detection in MALDI spectra and submission of peak lists to the database were done using Mascot wizard (Matrix Science). Peptide mass tolerance was set to 50 ppm with carbamidomethyl-cystein set as fixed modification, oxidation of methionine as variable modification and 1 missed cleavage site allowed.
3. RESULTS
Intracellular replication of M. bovis BCG within murine peritoneal macrophages was assessed by measuring the [3H] thymidine uptake. Table 1 shows four-fold increase in thymidine uptake in test group which was maximal when macrophage to mycobacterium ratio was 1:10. It was observed that after this infection ratio there was decrease in thymidine uptake with increasing infection ratio, which could be due to clumping of M. bovis BCG. These observations suggested that this in vitro infection model was able to support the growth of M. bovis BCG.
The most striking difference observed by SDS-PAGE was the induction of some proteins in infected cell lysate as compared to those of mycobacterial or macrophage cell lysates (Figure 1). Seven proteins were differentially expressed during macrophage-mycobacterium interaction and these proteins were further identified by MALDI-TOF (Table 2). Spot 1 (Mb2271) was identified as probable propionyl-CoA carboxylase beta chain 6 which is important for cell envelope lipid biosynthesis. Spot 2 (Mb3761c) encodes UPF0089 protein which is basically uncharacterized protein family. Spot 3 (Rv00 46c/MT0052) encodes a hypothetical protein and could not be assigned any function. Spot 4 was identified as

Table 1. [3H] Thymidine Incorporation to Mycobacterium bovis BCG. The values are the mean cpm (counts per minute). The SEM of triplicate cultures was < 12% of the mean.
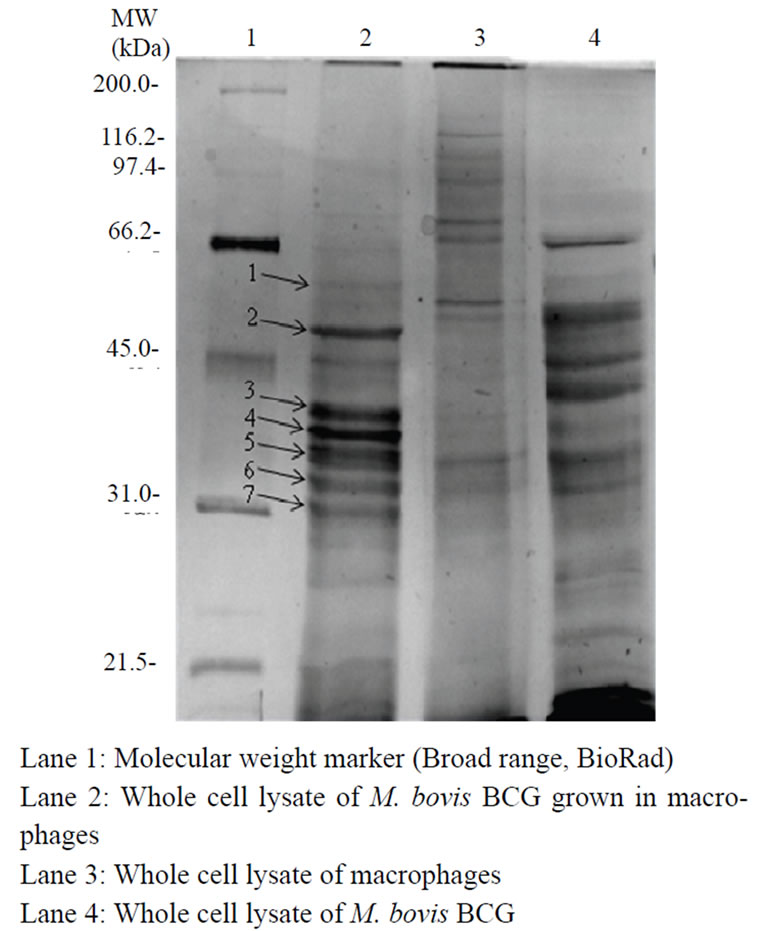
Figure 1. SDS-PAGE profile showing proteins expressed during macrophage-mycobacterium interaction.
riboflavin biosynthesis protein ribAB with accession number Mb1450. Spot 5 with accession number Mb22 60c was identified as cobalamin biosynthesis protein cobD. Spot 6 (Mb2729) is a transcription initiation factor sigma B and is suggested to auto-amplify its own expression under certain stress conditions. Finally, spot 7 (Mb3581) was identified as putative CoA-transferase subunit alpha and has a role in lipid metabolism. Experiments were repeated three times. These proteins might play some role in intracellular survival of mycobacteria within the macrophages.
4. DISCUSSION
The present study deals with development of an in vitro infection model system to study protein profile during macrophage-mycobacterium interaction. Model systems have proved of central importance in determining the activities associated with mammalian cells. To explore the strategies employed by mycobacteria to survive within the macrophages, studies have mostly been carried out with cell lines [3,15]. Our study, however, varies with respect to host macrophage system as we have used natural macrophages from murine peritoneum. Mice are the most widely used vertebrates with short reproductive cycle, short life span, small size and low cost of maintenance making them suitable substitutes in biomedical research.
Our results with thymidine uptake assay clearly suggest that the present in vitro infection model was able to support the growth of M. bovis BCG in murine macrophages. The pattern of cell lysate proteins indicate that adjustments in the pattern of gene expression do occur during macrophage-mycobacterium interaction. The present study revealed seven proteins that were upregulated during macrophage-mycobacterium interaction. Four of the identified proteins (propionyl-CoA carboxylase beta chain 6, riboflavin biosynthesis protein ribAB, Cobalamin biosynthesis protein cobD and putative CoA-transferase subunit alpha) were found to belong to the lipid metabolism category. Propionyl-CoA carboxylase catalyses the carboxylation reaction of propionyl CoA and methylmalonyl CoA is formed. Propionyl CoA is a key precursor in several lipid biosynthetic pathways in M. tuberculosis [16]; however, accumulation of propionate is toxic to the cell and efficient mechanisms are required for its disposal [17]. This dual nature implies a central role for propionate metabolism in the growth and persistence of M. tuberculosis in vivo [18]. Savvi et al. [19] demonstrated the ability of M.tuberculosis to utilize propionate as the sole carbon source in absence of functional methylcitrate cycle provided cobalamin (vitamin B12) is supplied exogenously and thus reinforces the potential relevance of vitamin B12 to mycobacterial patho

Table 2. Details of upregulated proteins identified by mass spectrometry.
genesis. Thus the differential expression of cobalamin biosynthesis protein in our experiment is significant. Riboflavin (vitamin B2) biosynthesis has been reported to be essential for in vivo survival of a number of bacterial species because of scarcity of riboflavin in mammalian cells [20]. Riboflavin is the precursor of the coenzymes flavin mononucleotide phosphate and flavin adenine dinucleotide phosphate that are essential compounds for basic metabolism. CoA transferases catalyze the reversible transfer of CoA from CoA-thioesters to free acids and are involved in lipid biosynthesis. Increased expression of these proteins directly or indirectly support their probable role during growth of mycobacteria within macrophages.
Transcription initiation factor sigma B is induced under various stress conditions like heat shock, cold shock, low aeration and limited or no substrate as seen in stationary phase [21,22]. Upregulation of Sigma factor B during macrophage-mycobacterium interaction suggests its role in intracellular survival. Earlier reports have revealed the upregulation of stress proteins under oxidative stress [23] and upon infection of macrophage [3]. No information however is available for the hypothetical protein and UPF0089 protein. Our findings indicate that M. bovis BCG mounts a specific response to the host intracellular environment and the expression of these preferred proteins plays an important role in its intracellular survival.
It is rather unlikely that only expression of seven proteins is sufficient for intracellular survival. Probable reasons for not finding more proteins could be due to technical reasons. Future studies would employ approaches like application of more sensitive and complementary proteome techniques or analyzing at a point where protein profile of mycobacteria is quite stable. Analysis of proteins by two-dimensional gel electrophoresis could not be carried out due to difficulty in recovery of sufficient amount of intracellular BCG proteins. MS identification of mycobacterial proteins from two-dimensional gels has mainly been applied to mycobacteria growing in broth grown cultures where abundant amounts of protein are available for analysis. Only a few studies have utilized the classical proteomic approach to study the mycobacterium-macrophage interactions due to the paucity in recovery of intracellular mycobacteria using infection models [8]. The present study is an attempt in the latter approach.
To summarize, our results clearly indicate that this in vitro infection system is an ideal model system to characterize the differentially expressed genes during the interaction of mycobacterium with macrophages. The employment of such model systems will help in understanding the early molecular events during mycobacterium-macrophage interactions, and thus mycobacterial virulence and pathogenicity. It is hoped that such proteomic studies may contribute significantly to understand mycobacterial pathogenesis and may help in the development of new drug targets and attenuated vaccines.
5. ACKNOWLEDGEMENTS
The authors are grateful to Dr. VM Katoch & Dr. UD Gupta, for their help. We also thank Mr. Ajeet Pratap Singh for technical assistance. This work was supported by grant from DST, New Delhi (No. SR/FTP/LS-A-80/ 2001). NS is SRF (CSIR-UGC). PS and BK are SRFs, ICMR.
6. DISCLOSURES
This paper is unique and is not under consideration by any other publication and has not been published elsewhere. This manuscript has been read and approved by all authors. The authors report no conflicts of interest.