Structural Study of Methylated and Non-Methylated Duplexes by IR Fourier Spectroscopy ()
1. Introduction
CpG islands in native DNA are known to exist mainly in the methylated form [1] . The methylation of DNA controls all genetic processes and serves as the mechanism of cell differentiation and gene repression [2] [3] . In some works, (CCGСС)n duplexes were studied by IR Fourier spectroscopy. Consecutive methylation of the duplex produced determinate changes in the region of 820 - 840 cm−1. The observed bands were attributed to the B-structure and BI→BII structural transitions [4] . On the other hand, investigation of molecular dynamics in the methylated (ССGСС)n duplex predicted the appearance of B→Z transition [5] . Quite interesting is the IR Fourier spectroscopy study of (ССGСС)n duplex in dependence on temperature in the range of 200 - 290 K. The B→Z and ZI→ZII transitions were revealed in the indicated temperature region [6] . Methylated (ССGСС)n duplexes are certainly of great interest [2] [3] . However, we think that investigation of the minimally methylated duplexes with a single ССGСС site in the AT enriched nucleotide environment is also important. The model system was represented by oligo-deoxyribonucleotide duplex consisting of 23 pairs of bases, which had a single (GCNGC) site simulating the binding site of THC-apoA-I complex on native DNA and oligonucleotides [7] - [9] . Prior to examining the interaction of methylated duplex with THC-apoA-I complex, we studied the structure of the duplex by IR Fourier spectroscopy. The goal of the work was to investigate structural changes of methylated duplex using IR Fourier spectroscopy.
2. Materials and Methods
Deoxyribooligonucleotides:
I) 5’-GAGTTTAGCGGCTATCGATCTCT- 3’ and II) 5’-AGAGATCGATAGCCGCTAAACTC- 3’ were syn- thesized at the Institute of Chemical Biology and Fundamental Medicine SB RAS (Novosibirsk).
The [γ-32P] ATP tracer (Isotope, St. Petersburg, 1 Ci/mol) was introduced to the 5’-end of oligonucleotides using polynucleotide kinase of T4 bacteriophage (SibEnzyme, Novosibirsk).
The OLI/OLII duplex was obtained by annealing of initial oligonucleotides taken in equimolar ratio (90˚C, 3 min, gradual cooling to room temperature).
Methylation of the duplex catalyzed by М. Fsp4HI methyltransferase (SibEnzyme, Novosibirsk) was carried out according to the analytical certificate of the enzyme: 1.7 × 10−8 M of the OLI/OLII duplex, 80 mM of SAM, 1 a.u. of methyltransferase М. Fsp4HI in a methylation buffer (10 mM Tris-HCl, pH 7.6, 10 mM MgCl2, 1 mM DTT). The mixture was incubated at 30˚C for 1 h. Methylation of the duplex involved cytosine from each nucleotide sequence to obtain one molecule of methylated cytosineper one oligonucleotide sequence. The reaction was stopped by adding 1 ml of 0.5 m EDTA [10] .
IR Fourier Spectra. Spectra of the duplexes were recorded in a Fourier spectrometer (Nicolet 6700, Thermo Scientific) using the Frustrated Total Internal Reflection (FTIR) method with a diamond cell at a resolution of 4 cm−1 in the frequency range of 900 - 4000 cm−1, in isotonic Tris buffer with the addition of K, Na salts (pH 7.4). Volume of the tested solution was 6.0 ml. A sample was covered with a fluoroplastic cap, which was pressed to the cell. The optical chamber was sealed and purged with dry air. The time of spectra recording ranged from 30 s to 1 min. Mathematical treatment of the spectra was performed using a special OMNIC software attached to the spectrometer.
3. Results
IR Fourier spectra. Analysis of the spectra revealed the absorption bands at 1085.67 (the PO2 bond) and 1044.44 (the C4-О4-С5-О5 bond) [8] [9] for the duplex (dupl_23) and 1112.56 and 1044.31 cm−1 bands for the methylated duplex (duplm_23) (Figure 1(a) and Figure 2(a)). It is seen that the absorption band of the PO2 bond in duplm_23 is shifted to the short-wave region by 26.89 cm−1. In addition, the integrated intensity ratio S1044.44/S1085.67 was equal to 4.38 in the duplex, while in duplm_23 the S1044.31/S1112.56 ratio was 2.82 (Table 1). Thus, the first value is greater than the second one by a factor of 1.55. A substantial shift of the absorption band of PO2 bond to the short-wave region and changes in the ratio of the indicated bands testify to the conformational changes in the backbone that occur due to methylation of bases (cytosine). The methylation involved only two bases residing in different oligonucleotide chains, while the backbone changed only slightly. This suggests the effect of hyperconjugation of the methyl group with the base [11] , which produces radical changes in the electron distribution and order of the bonds in bases and side radicals, and hence leads to conformational changes in the backbone and the entire molecule. The absorption bands at 1199.17 and 1218.25 cm−1 (dupl_23) as well as 1213.03 and 1230.93 cm−1 (duplm_23) can be attributed to the P=O bonds [8] [9] (Figure 1 and Figure 2). In this case, a 12 - 14 cm−1 shift to the short-wave region is also observed. The indicated changes suggest that the pitch of a helix decreases due to an increased ordering of bases and a stronger Van der Waals interaction between them. In this case, of special interest in the analysis of CH bonds. In dupl_23, deformation vibrations of
![]() (a)
(a)![]() (b)
(b)
Figure 1. (a) FTIR spectra of dupl.: (Сtris-HClbuf = 0.01 M, pH 7.4); ν = 800 - 1300 cm−1; (b) FTIR spectra of dupl.: (Сtris-HClbuf = 0.01 M, pH 7.4); ν = 2700 - 3100 cm−1.
![]()
Table 1. Frequency characteristics of the FTIR spectra of methylated (duplm) and non-methylated (dupl) duplexes.
*Indicates band splitting.
![]() (a)
(a)![]() (b)
(b)
Figure 2. (a) FTIR spectra of duplm: (Сtris-HClbuf = 0.01 M, pH 7.4); ν = 800 - 1300 cm−1; (b) FTIR spectra of duplm: (Сtris-HClbuf = 0.01 M, pH 7,4); ν = 2700 - 3100 cm−1.
the CH bonds show up as the absorption bands at 1418.58 and 1453.88 cm−1. Whereas in duplm_23 these bands (1419.50 and 1459.24 cm−1) are split. In addition, quite intense absorption bands at 993.71 and 922.68 cm−1 (the CC skeletal vibrations) are recorded in duplm_23 (Figure 1 and Figure 2), while in dupl_23 these bands are absent; note that the band at 922.68 cm−1 is assigned to the Z conformation.
CH stretching vibrations. Analysis of the spectra in the short-wave region revealed the presence of some absorption bands for dupl_23, in particular, 3076.47, 2979.64, 2944.32 and 2901.99 cm−1. The bands at 3063.35, 2947.97, 2936.13 and 2889.90 cm−1 were recorded for duplm_23 (Figure 1(b) and Figure 2(b)). It is seen that all the absorption bands in duplm_23 are shifted with respect to same bands indupl_23; the band at 2979.64 is shifted by 32.33 cm−1 to the long-wave region. This substantial shift can be attributed to an increased ordering of bases and monosaccharides and stronger Van der Waals forces between them.
Thus, the analysis of IR Fourier spectra of dupl_23 and duplm_23 showed that their conformations differ, duplm having a more ordered structure than the duplex. This is indicated by the frequency shift in the backbone and CH bonds region as well as by changes in the ratios of the absorption bands corresponding to the backbone and changes in intensity of the bands corresponding to CH stretching vibrations. Changes in the integrated intensity ratio S1044/S1085 for duplm testifies to the order→order structural transition that occurs due to methylation of the duplex.
![]()
Table 2. Absoption bands which correspond Z-structure [6] .
4. Discussion
The study of dupl and duplm demonstrated that the integrated intensity ratio of the absorption bands S1044/S1085 was equal to 4.4 in dupl and 2.8 in duplm. This 1.5-fold difference indicates that integrated intensity of the absorption band at 1085 cm−1 strongly decreases after methylation of the duplex. The band at 1085 cm−1 belongs to the 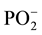 group (symmetric vibrations).
group (symmetric vibrations).
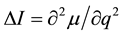 , [12]
, [12]
where μ is the dipole moment and q is the generalized coordinate. As follows from this expression, the dipole moment of the 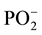 group decreases, i.e. the duplex becomes more hydrophobic after the methylation. In addition, the duplm backbone is described by a quartet (the absorption bands at 1112, 1083, 1043 and 994.5), and the dupl backbone is described by a triplet (1085, 1044.4 and 1001.3 cm−1). What is the reason for decreasing the dipole moment in the phosphate group? In our opinion, this is related to manifestation of the electron-donor properties of methylated bases as a result of hyperconjugation [11] , which leads to partial neutralization of the positive charge of phosphorus atom and enhances the hydrophobic properties of duplm.
group decreases, i.e. the duplex becomes more hydrophobic after the methylation. In addition, the duplm backbone is described by a quartet (the absorption bands at 1112, 1083, 1043 and 994.5), and the dupl backbone is described by a triplet (1085, 1044.4 and 1001.3 cm−1). What is the reason for decreasing the dipole moment in the phosphate group? In our opinion, this is related to manifestation of the electron-donor properties of methylated bases as a result of hyperconjugation [11] , which leads to partial neutralization of the positive charge of phosphorus atom and enhances the hydrophobic properties of duplm.
Essential changes occur also in the region of CH stretching vibrations. A quartet (the absorption bands at 2979, 2944, 2918 and 2901 cm−1) was observed for dupl, while only two main bands (at 2945.7 and 2888.4 cm−1) were seen in this region for duplm. The 2979→2945.7 cm−1 shift is equal to 33.3 cm−1; this is a pronounced shift that can be interpreted as a structural transition from one conformation to another. The main reason is related to a considerable ordering of bases, formation of the energetically more favorable conformation of deoxyribose, and their spatial ordering with respect to the duplex axis, which produces changes in the backbone structure, in particular, the endo 2→endo 3 conformational changes of deoxyribose. Exactly these conformations radically change the structure of DNA backbone [13] [14] . We think that after methylation of the duplex, individual bases become highly cooperative and form a joint “rigid block”.
It should be noted that methylation is accompanied by the order→order structural transition and, in particular, by the В→Z transition. Methylation induces conformational changes and decreases enthalpy of the transition [5] . Thus, we think that the changes observed in the region of backbone and CH stretching vibrations are caused by the В→Z transition. This is indicated also by the absorption bands revealed in methylated duplexes, which correspond to the Z structure (Table 2).