Microbial Observation in Bioaccumulation of Heavy Metals from the Ash Dyke of Thermal Power Plants of Chhattisgarh, India ()
1. Introduction
Global pollution is increasing due to the variations in natural and anthropogenic activities, leading to contaminations in various aquatic and terrestrial ecosystems with heavy metals, organic and inorganic chemical compounds. Among various pollutants, heavy metals are released into soils [1] . Presence of heavy metals such as Cu, Ni, Cr, and Cd, in fly ash causes metal toxicity in plants [2] [3] . Toxic metals are common contaminants of natural waters and may adversely affect the potentially important biodegradation processes naturally occurring in environment. Sources of these pollutants may include leachates from hazardous waste sites, discharges from industrial plants and effluents from wastewater treatment plants, manufacturing of alkaline storage batteries and combustion of fossil fuels [4] . The metal pollution is of great concerns, as these hazardous pollutants are accumulated in living organisms including microorganisms, plants, animals, and human [5] -[8] and are responsible for various metabolic and physiological disorders [9] [10] . The ideal solution of the abatement of pollution for these non-biodegradable metals is “bioremediation” which is found to be an efficient strategy to manage and recover the contaminants from the environment [11] . Microbial diversity is the most extraordinary resource of the life in the biosphere which we have just started to understand and explore. Some of the metals such as zinc, copper, nickel and chromium are indispensible or useful micronutrient for the growth and development of organisms [12] . Metals like cadmium, mercury and lead have no physiological effect instead show deleterious impact when present above threshold level by Gadd [13] . Carboxylic groups present in the techoic acid, associated to the peptidoglycan layer of the gram positive bacteria are found to be responsible for higher metal uptake as observed by Issazadeh et al. [14] . Costa et al. [15] observed and concluded that the bioaccumulation of Cu, Zn, Cd and Pb by Bacillus sp., Bacillus cereus, Bacillus sphaericus and Bacillus subtilis and the best results obtained with Bacillus subtilis and Bacillus cereus.
2. Materials and Methods
2.1. Sampling
Soil and effluent samples were obtained from Ash Dyke area of four Thermal Power Plants located at Bharat Aluminium Company (BALCO), Chhattisgarh State Electricity Board (CSEB), Korba, Thermal Power Cooperation (NTPC), Bilaspur and KSK Akaltara, Chhattisgarh. The samples were collected in sterilized flasks and beakers and after sealing with parafilm, they were stored at 4˚C in the fridge.
2.2. Isolation of Microorganisms
Different bacteria were isolated from the Ash Dyke area of four different plants situated in Chhattisgarh, using serial dilution technique in nutrient medium. Since nutrient medium is the most familiar medium to all microbiologists and consulting articles of previous works [16] -[18] , and it consists of all the basic nutrients required for the growth of the microorganisms, it is selected as the best medium for growth. 1 g of soil and 1 ml of effluent was poured into different flasks having 9 ml of saline. From these samples then, their respective serial dilution series was prepared. Out of which, 10−3 and 10−4 dilutions were selected for soil and were spreaded onto the sterilized petriplates containing the nutrient medium [19] [20] . Isolates were named as SM1 to SM25. Then the 25 possibly different isolates were selected randomly from the plates and were streaked by pick and patch method onto the separate Petri plates containing Pb, Hg, Cu, Co, Ni and Mn in concentration 0.08 mM/ml.
2.3. Storage of Pure Cultures
Constant subcultures of the isolates are maintained in EG medium at regular intervals. The pure cultures are preserved in 10% glycerol + EG medium at −70˚C in Deep freezer.
2.4. pH and Temperature
Selection of optimal pH and temperature for growth is done by selecting; pH 4, 6, 8, 10 and 4˚C, 15˚C, 29˚C, 35˚C for different pH respectively. Both tests were performed with metal containing medium and it was found that pH 6 and temperature 35˚C was most suited for the isolates for sustaining their growth.
2.5. Bacterial Minimum Inhibitory Concentration (MIC) and Maximum Tolerance Concentration (MTC)
The minimum inhibitory concentration (MIC) has formed the basis for the selection of the potent isolates and henceforth 25 bacteria were found out of 150 which could grow at 0.08 mM/ml concentration of the metals. But only 3 among 25 could grow at 0.2 mM/ml concentration, so these three were selected for further studies. MTC of these selected isolates towards selected heavy metal has been checked in 0.2, 0.4 and 0.6 mM/ml of the six metals in nutrient agar medium (NAM). Following component in the NAM were used i.e. Peptone 5.0, NaCl 5.0, Beef extract 1.5, Yeast extract 1.5, pH 7.4 ± 2. Media grade used was of HiMedia.
2.6. Identification of the Isolates
Biochemical tests formed the basis for the detection of the three isolates. Bacterial isolates were identified as per the standard methods following Bergeys Manual of Determinative Bacteriology.
2.7. Biosorption of Metals by the Bacteria
Metals were added to EG broth medium at concentration of 0.2, 0.4 and 0.6 mM/ml and the 6 isolates were separately inoculated. The flasks were kept at 35˚C for 48 hrs at 120 rpm. Then the cells were harvested by centrifugation and suspended in 1 ml distilled water. The metal content of bacterial cells is determined after acid dissolution of bacterial cells using the method of Ganje and Page [21] metal concentration was measured by AAS.
2.8. 16S rDNA Sequencing
Genomic DNA was isolated from the sample. The ~1.5 kb 16s-rDNA fragment was amplified using high-fidel- ity PCR polymerase. The PCR product was sequenced bi-directionally. The sequence data was aligned and analyzed to identify the bacterium and its closest neighbors. The primers used are, 16 s Forward Primer: 5’- AGHGTBTGHTCMTGNCTCAS-3’. 16 s Reverse Primer: 5’-TRCGGYTMCCTTGTWHCGACTH-3’. The Ther- mal Cylcer: ABI2720 was used for this process [22] .
3. Results
The present result will give the details of Isolation and 16S rDNA based characterization through cluster analysis and Biosorption of Metals on the cell increasing and decreasing trends in various concentrations by the Bacteria.
3.1. Enumeration, Isolation and 16S rDNA Based Characterization
In the present study the total viable count appeared to be 7.8 × 106 cfu/g average of the four samples selected, but varied from 3.5 × 106 to 5.5 × 106 cfu/g on the multiple-metal supplemented plate (Table 1). Out of 150 isolates, 25 were selected to grow in 0.08 mM/ml concentration. A total of 25 isolates that varied in shape, color and morphology were selected randomly from the multi-metal supplemented plate (0.08 mM/ml concentration). But when concentration further increased to 0.2 mM/ml of different metals, only selected Bacillus group could survive and were found to grow till 0.6 mM/ml concentration. The entire practical’s carried out in triplicates to check the reliability of the results. After biochemical identification and 16S rDNA based identification the useful isolates were determined as Bacillus cereus (SM2, SM3), and Bacillus subtilis (SM12) as determined and phylogenetic tree were plotted by cluster analysis [23] . Table 2, representing the Bioaccumulation (mg/g dry weight of the cells) of six heavy metals with different concentration by SM2, SM3 and SM12 after 24 hrs. Phylogenic analysis of isolation and identification of bacteria were done by cluster analysis as phylogenetic tree (Figures 1-3).
3.2. Biosorption of Metals by the Bacteria
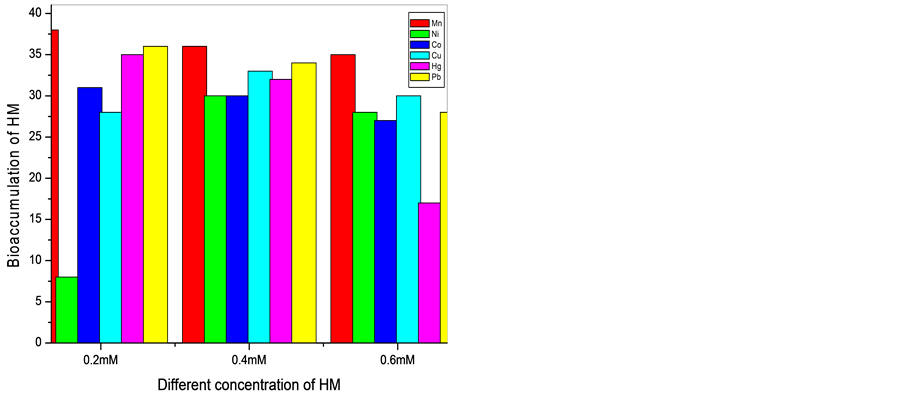
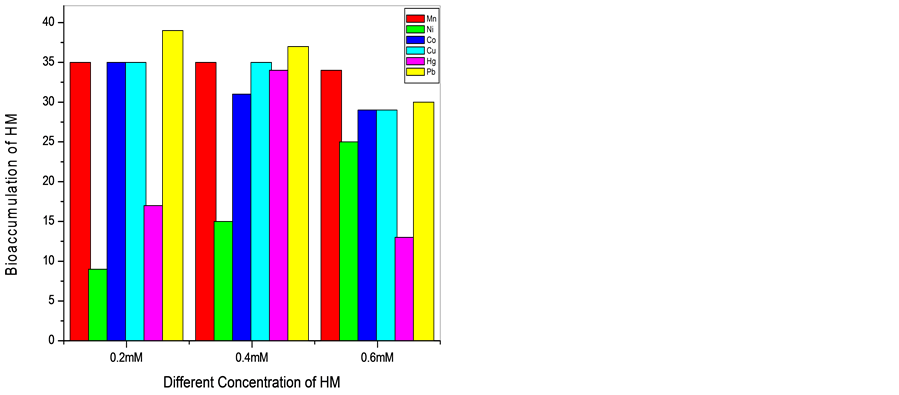
It is observed that the bioaccumulation of the metals on the cells decreased when concentration increased from 0.2 to 0.6 mM/ml of the media. The Mn resistant bacterium (SM2, SM3, and SM12) accumulated Mn around 5 - 39 mg/g at concentration range 0.2 to 0.6 mM/ml of the media. Ni resistant bacteria (SM2, SM3, and SM12) could accumulate 8, 9, 34 and 32 mg/g dry weight of the cells when concentration was 0.2 mM/ml of the media. But at concentration 0.4 and 0.6 mM only SM2 and SM3 could accumulate Ni at range of 15 to 30 mg/g of the dry weight of the cells. When the isolates were grown in 0.2, 0.4 and 0.4 mM Co, accumulation varied from 6 - 35 mM/ml of the media. The Cu resistant cells could accumulate 6 - 35 mg/g at the three mentioned concentrations
![]()
Table 1. Bacterial count on heavy metal supplemented media (CFU/g).
![]()
Table 2. Bioaccumulation (mg/g dry wt. of the cells) of six heavy metals (0.2, 0.4 and 0.6 mM/ml concentration) by SM2, SM3 and SM12 after 24 hrs.
of the metal. Hg accumulation in the cells varied from 10 - 35 mg/g dry weight of the cells. Pb found to be at the range of 10 - 39 mg/g of the dry weight of the cells (Graph 1 and Graph 2).
4. Discussion
In the present study, three Bacilli isolated from the ash sample of power plants were selected owing to their maximum relative growth and metal accumulation ability. These results are consistent with the screening for multi-metal resistant bacteria performed in previous studies [24] [25] . While working on a lead resistant Bacillus strain isolated from a slag disposal site, Pandey et al., [26] found that capability of efficiently accumulating lead.
(a) (b)
Graph 1. Representing the bioaccumulation (mg/g dry wt. of the cells) of six heavy metals (HM) with 0.2, 0.4 and 0.6 mM/ml concentration by SM2 and SM3 respectively after 24 hrs.
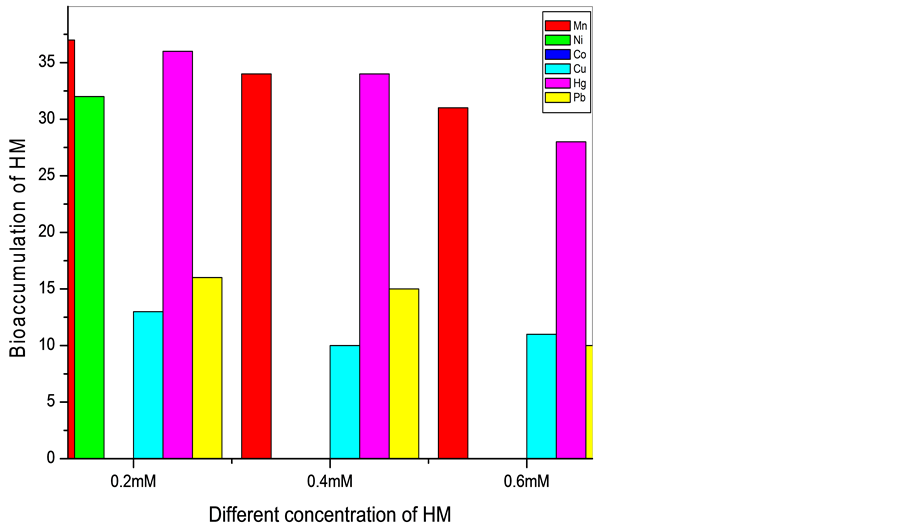
Graph 2. Representing the bioaccumulation (mg/g dry wt. of the cells) of six heavy metals (HM) with 0.2, 0.4 and 0.6 mM/ml concentration by SM12 after 24 hrs.
Similar studies carried out by Singh et al., [27] on protease producing halotolerant Bacillus cereus SIU1 strain isolated from a non-saline environment was also found to be resistant to heavy metals (As, Pb and Cs). In metals, Cd, Hg, and Pb have no known physiological functions [13] . However, the higher concentration of these metals above threshold levels has deleterious impact on the microbial communities and their functional activities in soils. Otherwise, microorganisms exposed to the higher concentrations of toxic heavy metals may develop resistance against the elevated levels of these metals [28] .
In addition, microorganisms inhabiting in metal polluted soils have evolved various methods to resist themselves against metal stress. One of the microbial processes as a bioremediation tool to remove heavy metals from soils is bioaccumulation [29] which is an active process dependent upon metabolic energy of microorganisms by Rani & Goel [9] . The result clearly illustrated that the accumulation of the metals on the cells of Bacillus cereus and Bacillus subtilis decreased with increased concentrations of the metals in the media. The results are in correlation with the work done by Murthy et al. [30] .
5. Conclusion
The present experiment of metal accumulation by the metal resistant isolates from the ash dyke sample revealed a positive indication for the use of the bacterial isolates in metal contaminated soils for their bioremediation. Although our study was carried out under idealized laboratory conditions, results suggested that some general considerations could be implied for these isolates to be used under field conditions.