Regularization and Estimation in Regression with Cluster Variables ()
1. Introduction
We are often interested in finding important variables that are significantly related to the response variable and can be used to predict quantities of interest in regressions and classification problems. Important variables are often shown in clusters where variables in the same cluster are highly correlated and have similar pattern relating to the response variable. For example, a major application of microarray technology is to discover important genes and pathways that are related to clinical outcomes such as the diagnosis of a certain cancer. Typically, only a small proportion of genes from a huge bank have significant influence on the clinical outcome of interest. In addition, expression data frequently have cluster structures: the genes within a cluster often share the same pathway and are therefore similarly related to the outcome. When regression is adapted in this setting, we often face the challenge from multi-collinearity of covariates. An ideal variable selection procedure should be able to find all genes of important clusters rather than just some representative genes from the clusters. Typically, two characteristics, pointed out by [1] , evaluate the quality of a fitted model: accuracy of prediction on new data and interpretation of the model. For the latter, the sparse model with fewer selected covariates is preferred for interpretation due to its simplicity. However, when multiple variables share the same mechanism for explaining the response, all the involved variables should have an equal chance of being selected, and should exhibit the same relationship to the outcome in the fitted model, for scientific reasoning.
It is well known that the ordinary least square estimate (OLS) in linear regression often performs poorly when some of the predictors are highly correlated. OLS would generate unstable results where the estimates have inflated variances. Regularizations have been proposed to improve OLS. For example, ridge regression [2] penalizes the model complexity by the 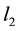 penalty of the coefficients. This method was proposed to solve the collinearity problem by adding a constant to the diagonal terms of
penalty of the coefficients. This method was proposed to solve the collinearity problem by adding a constant to the diagonal terms of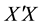 , where
, where 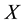 is the observation or design matrix. Ridge regression stabilizes the estimates through the bias-variance trade-off. It can often improve the predictions but cannot select variables. [3] proposed the Lasso method by imposing an
is the observation or design matrix. Ridge regression stabilizes the estimates through the bias-variance trade-off. It can often improve the predictions but cannot select variables. [3] proposed the Lasso method by imposing an 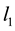 -penalty on the regre- ssion coefficients. Lasso is a promising method, as it can improve prediction and produce sparse models simultaneously. However, when high correlations among predictors are present, the predictive performance of Lasso is dominated by ridge regression [3] . Moreover, when there is a cluster of variables, in which each variable associates with the response variable similarly, Lasso tends to arbitrarily select one variable from the cluster instead of identifying the cluster [1] ; see also Section 2 for more discussion. Elastic Net, proposed by [1] , combines both
-penalty on the regre- ssion coefficients. Lasso is a promising method, as it can improve prediction and produce sparse models simultaneously. However, when high correlations among predictors are present, the predictive performance of Lasso is dominated by ridge regression [3] . Moreover, when there is a cluster of variables, in which each variable associates with the response variable similarly, Lasso tends to arbitrarily select one variable from the cluster instead of identifying the cluster [1] ; see also Section 2 for more discussion. Elastic Net, proposed by [1] , combines both 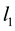 and
and 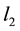 penalties of the coefficients as the regularization criterion. The method is promising in that it encourages cluster effects and shows improved predictive performance over Lasso. Elastic Net can automatically choose cluster variables and estimate parameters at the same time. Many other methods can be used to choose clustered variables, such as principal component analysis (PCA). [4] defined “eigen-arrays” and “eigen-genes” in this way. But PCA can not choose sparse models. [5] proposed sparse principal component analysis (SPCR), which formulated PCA as a regression-type optimization problem, and then obtained sparse loadings by imposing the Elastic Net constraint. SPCR can successfully yield exact zero loadings in principal components. However, for each principal component, a regularization parameter has to be selected, which results in an overwhelming computational burden when the number of parameters is large. Other penalized regression methods have been proposed for group effect [6] - [13] . However, these methods either pre-suppose a grouping structure or assume that each predictor in a group shares an identical regression coefficient.
penalties of the coefficients as the regularization criterion. The method is promising in that it encourages cluster effects and shows improved predictive performance over Lasso. Elastic Net can automatically choose cluster variables and estimate parameters at the same time. Many other methods can be used to choose clustered variables, such as principal component analysis (PCA). [4] defined “eigen-arrays” and “eigen-genes” in this way. But PCA can not choose sparse models. [5] proposed sparse principal component analysis (SPCR), which formulated PCA as a regression-type optimization problem, and then obtained sparse loadings by imposing the Elastic Net constraint. SPCR can successfully yield exact zero loadings in principal components. However, for each principal component, a regularization parameter has to be selected, which results in an overwhelming computational burden when the number of parameters is large. Other penalized regression methods have been proposed for group effect [6] - [13] . However, these methods either pre-suppose a grouping structure or assume that each predictor in a group shares an identical regression coefficient.
In practice, we often have some prior knowledge about the structure of variables and would like to make use of a priori information in analysis. For example, in gene analysis, we know the pathways and genes involved in these pathways. Therefore, we would like to group the involved variables in the same pathway together. Another example is in spatial analysis, we would like to keep a certain correlation structure among the spatial error terms. For example, sometimes we would like to fit a different coefficient for a certain variable at different regions (e.g., if the variable has different effect at different regions) but keep a correlation structure among the coefficients at neighborhood regions. The conditional autoregressive model (CAR, [14] ) is one of the methods that can be used to keep such correlation structure.
In this paper, we propose a method that encourages cluster variables to be selected together and can incorporate available prior information on coefficient structures in variable selection. When there is no prior information on coefficient structure, we propose a data augmentation algorithm to find the structure. Moreover, the method uses the Lasso regularization to choose sparse models. The proposed method can be solved by any efficient Lasso algorithm such as least angle regression (LARS, [15] ) and the coordinate-wise descent algorithm (CDA, [16] ). We call our method the Clustering Lasso (CL).
The rest of the paper is organized as follows. In Section 2, we review the Lasso method and discuss its limitation in identifying clustered variables. Then we propose the Clustering Lasso in a Bayesian setting. Its counterparts in the Frequentist setting and computational strategies are discussed in Section 3. Sections 4 and 5 demonstrate the predictive and explanatory performance of CL through real examples and simulations. Finally, conclusions and future work are discussed in Section 6.
2. Clustering Lasso in Bayesian Setting
Consider linear regression settings with the response vector 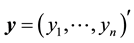 and
and 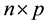 dimensional input matrix
dimensional input matrix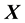 . The
. The 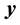 and columns of
and columns of 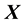 are centered and standardized to have the same
are centered and standardized to have the same 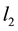 norm. The Lasso estimates
norm. The Lasso estimates 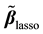 are calculated by minimizing
are calculated by minimizing
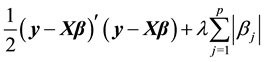 (1)
(1)
The solution of Lasso can be obtained through
LARS
or CDA. Compared with ordinary linear regressions, Lasso shows superior predictive performance and more stable estimates. Moreover, Lasso can often select variables and estimate coefficients simultaneously.
Group effect has been defined by [1] in the linear regression setting. Let 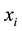 be the
be the 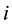 th predictor. The estimates of coefficients have the group effect if
th predictor. The estimates of coefficients have the group effect if ![]() would result in the estimated coefficients
would result in the estimated coefficients![]() . [1] further proved that if the solution for estimation is to minimize the objective function of the form:
. [1] further proved that if the solution for estimation is to minimize the objective function of the form:
![]() (2)
(2)
and the penalty term, ![]() , is strictly convex, then the estimates from Equation (2) enjoy the group effect property. In Lasso,
, is strictly convex, then the estimates from Equation (2) enjoy the group effect property. In Lasso, ![]() is
is ![]() norm of
norm of![]() , which is not strictly convex. Zou and
Hastie
proved that in this case Lasso estimates do not have the group effect. This is also understandable through the Lasso solution path from
LARS
. In
LARS
, suppose a variable
, which is not strictly convex. Zou and
Hastie
proved that in this case Lasso estimates do not have the group effect. This is also understandable through the Lasso solution path from
LARS
. In
LARS
, suppose a variable ![]() is selected in the model. Its coefficient solution path will move in a direction to reduce the correlation between
is selected in the model. Its coefficient solution path will move in a direction to reduce the correlation between ![]() and the current residual,
and the current residual, ![]() , until another variable, say
, until another variable, say![]() , has the same correlation to the current residual as does
, has the same correlation to the current residual as does![]() . At this point, variable
. At this point, variable ![]() is added into the model. If
is added into the model. If ![]() is highly correlated with
is highly correlated with![]() , when the correlation between
, when the correlation between ![]() and the residual decreases, so does that between
and the residual decreases, so does that between ![]() and the residual. Therefore, if
and the residual. Therefore, if ![]() has been included in the model, Lasso is less likely to select the highly correlated variable
has been included in the model, Lasso is less likely to select the highly correlated variable ![]() in the model. Consequently, Lasso cannot select clustered variables.
in the model. Consequently, Lasso cannot select clustered variables.
In a Bayesian setting, if ![]() is the ith row of
is the ith row of![]() , [3] showed that the Lasso solution is identical to the posterior mode of the coefficients when the prior distributions of the coefficients are set as independent double exponential distributions, where
, [3] showed that the Lasso solution is identical to the posterior mode of the coefficients when the prior distributions of the coefficients are set as independent double exponential distributions, where
![]()
In Lasso, the penalty term of model complexity is![]() . Because each coefficient is penalized equally, each one can be shrunk to zero independently. When the variables are clustered, an ideal solution path should be that the clustered variables are selected together. Therefore, we would like to penalize the coefficients with a restriction that keeps the correlation structure among the variables. With the penalization, if the coefficient of one variable is nonzero, those variables in the same cluster are less likely to be zero. For this purpose, we assume a correlation structure, specified as the structural correlation matrix
. Because each coefficient is penalized equally, each one can be shrunk to zero independently. When the variables are clustered, an ideal solution path should be that the clustered variables are selected together. Therefore, we would like to penalize the coefficients with a restriction that keeps the correlation structure among the variables. With the penalization, if the coefficient of one variable is nonzero, those variables in the same cluster are less likely to be zero. For this purpose, we assume a correlation structure, specified as the structural correlation matrix![]() , of the coefficients
, of the coefficients![]() .
.
For simplicity, assume that the variance of the random error, ![]()
![]() , and the structural correlation matrix
, and the structural correlation matrix ![]() are known. Then the likelihood and prior distributions can be set as:
are known. Then the likelihood and prior distributions can be set as:
![]()
Therefore, the posterior distribution of ![]() has the form
has the form
![]() (3)
(3)
with a vector![]() ,
,![]() . The posterior mode of
. The posterior mode of ![]() in the distribution (3) is the solution to
in the distribution (3) is the solution to
![]() (4)
(4)
Relating Equation (4) to the Bayesian Lasso solution to (1), we naturally infer the Clustering Lasso in Frequentist setting.
3. Clustering Lasso
3.1. Clustering Lasso and Its Grouping Effect
In Frequentist setting, we modify the penalization function in Lasso to retain a presumed correlation structure among coefficients. Let ![]() and
and ![]() be the jth element of
be the jth element of![]() . The Clustering Lasso estimate is defined as the solution to
. The Clustering Lasso estimate is defined as the solution to
![]() (5)
(5)
where ![]() is the regularization parameter. Note that instead of restricting
is the regularization parameter. Note that instead of restricting![]() , we restrict
, we restrict![]() . Therefore,
. Therefore,![]() ’s are not penalized independently and clustered variables could be chosen. Let
’s are not penalized independently and clustered variables could be chosen. Let![]() , with dimension
, with dimension![]() , be the jth row of
, be the jth row of ![]() and let
and let![]() , a
, a ![]() matrix. The penalty term used in expression (5) can also be written as
matrix. The penalty term used in expression (5) can also be written as![]() , which is intermediate between the
, which is intermediate between the ![]() penalty and the
penalty and the
![]() penalty. When
penalty. When ![]() is an identity matrix, the Clustering Lasso is identical to the ordinary Lasso method. Otherwise, the penalty function is strictly convex. Using Lemma 2 developed by [1] , the solution to Expression (5) has the group effect. Therefore, Clustering Lasso can select variables by clusters.
is an identity matrix, the Clustering Lasso is identical to the ordinary Lasso method. Otherwise, the penalty function is strictly convex. Using Lemma 2 developed by [1] , the solution to Expression (5) has the group effect. Therefore, Clustering Lasso can select variables by clusters.
Figure 1 illustrates the Clustering Lasso penalty contours with two predictors. The right figure shows the penalty contour when the two predictors are correlated and the left one shows the contour when the two predictors are independent, which is identical to the Lasso method. The sums of the squared errors have elliptical contours, centered and minimized at the full least squares estimate. The constraint region of Lasso is the diamond region![]() , while that for the Clustering Lasso is the parallelogram region defined by
, while that for the Clustering Lasso is the parallelogram region defined by
![]() . The optimal estimates are realized at the place where the elliptical contours first hit the
. The optimal estimates are realized at the place where the elliptical contours first hit the
constraint regions. The sides of the parallelogram are decided by the structural correlation matrix![]() .
.
![]()
Figure 1. Estimation picture for the Clustering Lasso when two predictors are independent (left, as lasso) and when two predictors are clustered (right).
3.2. Computation
The Clustering Lasso is an extension of the Lasso method. Let![]() . So the solution to Expression (5) is
. So the solution to Expression (5) is![]() , where
, where ![]() is
is
![]() (6)
(6)
Therefore, all the established algorithms for Lasso solution, such as the least angle regression (LARS, [15] ), could be used for Clustering Lasso.
3.3. The Clustering Lasso Algorithm
We can incorporate prior knowledge of clustering into a structural correlation matrix. For example, Kyoto Encyclopedia and
Genes
and Genomes (KEGG) and many other biological databases can be referred to in gene analysis to construct the structural correlation matrix. It is required that the structural correlation matrix be symmetric. When no prior information is readily adaptable, a natural method is to use the modified correlation matrix of the observed data, meaning that the coefficients should have a correlation structure that is similar to how the covariates are correlated. There are several well-established potential choices such as partial correlation matrix [17] . In this paper, we propose to use a modified correlation matrix so that if two variables ![]() and
and ![]() are not significantly correlated,
are not significantly correlated, ![]() , the ith row and jth column element of
, the ith row and jth column element of![]() , is set to be zero. As the solution
, is set to be zero. As the solution
for ![]() is
is![]() , zero elements in
, zero elements in ![]() are desired so that when
are desired so that when ![]() s are shrunk to zero, which is possible
s are shrunk to zero, which is possible
by the Lasso property, some ![]() s could also be shrunk to exact zero.
s could also be shrunk to exact zero.
In detail, we develop Algorithm 1―the Clustering Lasso algorithm. Let![]() ,
, ![]() , and
, and![]() , in
, in ![]() be three prespecified numbers, and
be three prespecified numbers, and ![]() be a
be a ![]() matrix.
matrix.
Algorithm 1 Clustering Lasso
1. For ![]()
for ![]()
do correlation test between ![]() and
and![]() , let
, let
![]()
2. Do
eigen
decomposition on ![]() so that
so that ![]() and let
and let ![]() if
if ![]() for
for ![]() .
.
3. Let ![]() and
and![]() .
.
4. Do Lasso on ![]() and get the coefficient solution
and get the coefficient solution![]() .
.
5. ![]() is the solution to Clustering Lasso.
is the solution to Clustering Lasso.
Note that only when some elements of ![]() are set to be zero, could
are set to be zero, could ![]() s be shrunk to exact zero when
s be shrunk to exact zero when![]() ’s are shrunk to zero by Lasso. A special case is when
’s are shrunk to zero by Lasso. A special case is when ![]() is a block diagonal matrix. To choose sparse models, we need to identify clusters of covariates, where variables in the same cluster are assumed to be correlated while those from different clusters are independent. For this purpose, there are two shrinkage steps in Algorithm 1. Step (1) shrinks the correlation coefficients to zero if there is no significant correlation between the pair of covariates at the significance level
is a block diagonal matrix. To choose sparse models, we need to identify clusters of covariates, where variables in the same cluster are assumed to be correlated while those from different clusters are independent. For this purpose, there are two shrinkage steps in Algorithm 1. Step (1) shrinks the correlation coefficients to zero if there is no significant correlation between the pair of covariates at the significance level ![]() or if the magnitude of correlation is smaller than a pre-set value
or if the magnitude of correlation is smaller than a pre-set value![]() . When two covariates are not correlated, there is little chance that the two variables relate to the response variable with the same underlying pathway. Therefore, the coefficients of the two variables can be estimated independently. Step (2) shrinks some eigen-values of
. When two covariates are not correlated, there is little chance that the two variables relate to the response variable with the same underlying pathway. Therefore, the coefficients of the two variables can be estimated independently. Step (2) shrinks some eigen-values of ![]() to zero if the corresponding eigenvector explains less than
to zero if the corresponding eigenvector explains less than ![]() times the total variance of
times the total variance of![]() . The two shrinkage steps cannot guarantee that some elements of
. The two shrinkage steps cannot guarantee that some elements of ![]() be zero. Subjective intervention can help for this purpose. One resolution is to cluster the covariates first and then calculate the correlation matrices for each cluster, which in turn used to build the diagonal blocks of
be zero. Subjective intervention can help for this purpose. One resolution is to cluster the covariates first and then calculate the correlation matrices for each cluster, which in turn used to build the diagonal blocks of![]() . In addition to building a diagonal block matrix, another resolution is to adapt shrinkage methods in the
eigen
decomposition process of
. In addition to building a diagonal block matrix, another resolution is to adapt shrinkage methods in the
eigen
decomposition process of![]() , so that some loadings of the eigenvectors might degenerate to 0. ScotLASS [18] and sparse principal component analysis (Zou et al., 2006) can serve this purpose. However, these methods require extra computations for each principal component, which brings in high computational costs. The nonzero elements of the jth row of
, so that some loadings of the eigenvectors might degenerate to 0. ScotLASS [18] and sparse principal component analysis (Zou et al., 2006) can serve this purpose. However, these methods require extra computations for each principal component, which brings in high computational costs. The nonzero elements of the jth row of ![]() imply that the corresponding covariates belong to the jth cluster. Ideally, their values should be proportional to the contributions of each covariate to the cluster in explaining the outcome. As pointed out by a referee of the paper, clusters in the proposed method are identified by rows of
imply that the corresponding covariates belong to the jth cluster. Ideally, their values should be proportional to the contributions of each covariate to the cluster in explaining the outcome. As pointed out by a referee of the paper, clusters in the proposed method are identified by rows of![]() , where
, where ![]() is defined as
is defined as ![]() with
with ![]() being the diagonal matrix of eigenvalues and
being the diagonal matrix of eigenvalues and ![]() columns of eigen- vectors of
columns of eigen- vectors of![]() . As in principal component analysis, the nonzero elements of
. As in principal component analysis, the nonzero elements of ![]() are difficult to interpret in practice. The referee recommends setting the elements of
are difficult to interpret in practice. The referee recommends setting the elements of ![]() to be 0 or 1 based on the absence or presence of non-zero elements, respectively. In the paper, we set the estimate of
to be 0 or 1 based on the absence or presence of non-zero elements, respectively. In the paper, we set the estimate of ![]() to be zero, if its estimated value is very close to zero, i.e. if
to be zero, if its estimated value is very close to zero, i.e. if![]() .
.
3.4. Choice of Tuning Parameters
Four parameters, ![]() , are to be chosen for Algorithm 1.
, are to be chosen for Algorithm 1. ![]() is the significance level used to decide whether the correlations between a pair of covariates should be considered to restrict the estimation of their coefficients. We usually select the significance level at 0.05, the traditional significance level. When the data set is large, we can reduce the significance level. Since the correlation would be always significant when little correlation exists and the number of observations is large, we set another restriction on the magnitude of correlation-
is the significance level used to decide whether the correlations between a pair of covariates should be considered to restrict the estimation of their coefficients. We usually select the significance level at 0.05, the traditional significance level. When the data set is large, we can reduce the significance level. Since the correlation would be always significant when little correlation exists and the number of observations is large, we set another restriction on the magnitude of correlation-![]() , above which we would like to use the correlation as a restriction to the coefficient parameters.
, above which we would like to use the correlation as a restriction to the coefficient parameters. ![]() is chosen subjectively by researchers. Algorithm 1 Step (2) is similar to the principal component analysis except that the
eigen
decomposition is based on the correlation matrix modified by Step (1).
is chosen subjectively by researchers. Algorithm 1 Step (2) is similar to the principal component analysis except that the
eigen
decomposition is based on the correlation matrix modified by Step (1). ![]() specifies the minimum proportion of variance explained by the
eigen
vector, below that, the
eigen
vector will not be used for further analysis.
specifies the minimum proportion of variance explained by the
eigen
vector, below that, the
eigen
vector will not be used for further analysis. ![]() is set at a small value, typically
is set at a small value, typically![]() , where
, where ![]() is the total number of covariates.
is the total number of covariates.
The last parameter to be tuned is![]() . In Lasso, the conventional tuning parameter is the fraction
. In Lasso, the conventional tuning parameter is the fraction ![]() of the
of the ![]() -norm. There are well-established methods for choosing
-norm. There are well-established methods for choosing![]() . Tenfold cross-validation (CV) on training data is the method we used in this paper. The training dataset is divided into ten folds randomly. One fold of the data is used as validation data, on which the prediction error is calculated based on the model fitted from the other nine folds of data.
. Tenfold cross-validation (CV) on training data is the method we used in this paper. The training dataset is divided into ten folds randomly. One fold of the data is used as validation data, on which the prediction error is calculated based on the model fitted from the other nine folds of data. ![]() is tested on a fine grid on
is tested on a fine grid on![]() . It takes the value that minimizes the averaged prediction error from CV. We can also use ten-fold CV to tune
. It takes the value that minimizes the averaged prediction error from CV. We can also use ten-fold CV to tune ![]() and
and![]() . We found that only a few representative values for
. We found that only a few representative values for
![]() need to be cross validated to obtain good results, which are
need to be cross validated to obtain good results, which are![]() .
.
4. Microarray Data Example
We used the proposed method on an Affymetrix gene expression dataset. The data were collected by Singh et al. [19] and consists of 12,600 genes, from 52 prostate cancer tumor samples and 50 normal prostate tissue samples. The goal is to construct a diagnostic rule based on the 12,600 gene expressions to predict the occurrence of prostate cancer. Support vector machine (SVM, [20] ), Ridge, Lasso, Elastic Net, Weighted Fusion (w.fusion) and Clustering Lasso were all applied to this dataset. We tried four types of Clustering Lasso methods:
1. CL1:![]() ,
, ![]() , and
, and![]() ;
;
2. CL2:![]() ,
, ![]() , and
, and![]() ;
;
3. CL3:![]() ,
, ![]() , and
, and![]() ;
;
4. CL4:![]() ,
, ![]() , and
, and![]() .
.
To apply these methods, we first coded the presence of prostate cancer as a 0-1 (no and yes) response![]() . The classification function is
. The classification function is ![]() (fitted value
(fitted value![]() ), where
), where ![]() is the indicator function. For comparison, we randomly select 52 samples as training data, based on which the diagnostic rules are constructed, and the rules are in turn tested on the remaining 50 samples.
is the indicator function. For comparison, we randomly select 52 samples as training data, based on which the diagnostic rules are constructed, and the rules are in turn tested on the remaining 50 samples.
The dataset was split 20 times. For each repetition, a 1000-gene set was preselected based on the training data to make the computation manageable. The genes are those “most significantly” related to the response, tested by individual t-statistics. Figure 2 shows the boxplots of the misclassification rates on the test data sets from different classifiers. The misclassification rates are summarized in Table 1. Overall, the misclassification rate from Clustering Lasso is competitive with Elastic Net and Ridge, and is better than Lasso, Weighted Fusion, and SVM. For the computational time, Clustering Lasso is comparable to the Lasso method and is much more efficient than Elastic Net and Weighted Fusion. Within the four Clustering Lasso methods, the ones with more restrictions on eigenvalues and the magnitudes of correlations perform a little bit worse.
Table 2 shows the average number of genes selected from the 20 repetitions based on different methods. The analyses were based on 1000 genes and 52 observations. We see that Lasso selected fewer than 52 genes. Elastic Net eliminated few genes―the average number of selected genes was close to 1000. Cluster Lasso identified about 25% genes as important. However, we do not know whether the chosen genes are, in fact, important or not. The efficiency of variable selection is further assessed by simulation studies.
![]()
Figure 2. Misclassification rates on singh data. ELAS stands for Elastic Net.
![]()
Table 1. Summary of Misclassification Rates on Singh data.
![]()
Table 2. Average number of genes selected by each method.
5. Simulation Studies
We applied Clustering Lasso on some simulations to test its prediction accuracy in regressions when compared with Ridge, Lasso, Elastic Net, and Weighted Fusion. The first three simulations are adapted from the Elastic Net paper [1] . To begin, datasets are simulated from the true model:
![]()
For each scenario, we simulated 100 data sets, each consisting of a training data set and an independent test data set. Here are the details of the four scenarios.
1. In example one, we simulated ![]() observations as training data and 200 observations as test data. We let
observations as training data and 200 observations as test data. We let ![]() and
and![]() . The pairwise correlation between
. The pairwise correlation between ![]() and
and ![]()
was set to be![]() .
.
2. In Example two, we simulated 200 training data and 400 testing data. There are 40 predictors such that
![]()
3. Example 3 has the group setting that ![]() and
and![]() , where the predictors are gene- rated as
, where the predictors are gene- rated as
![]()
As explained by [1] , three groups are equally important groups, and each group contains five covariates. We created ![]() observations as training data and 400 as testing data.
observations as training data and 400 as testing data.
The fourth simulation is a modification of the third example to emphasize the group effects. The true model has the form ![]() where
where![]() . The predictors we observed are
. The predictors we observed are
![]()
The latent variables, ![]() and
and![]() , directly relate to the response variable, where
, directly relate to the response variable, where ![]() is more important than
is more important than![]() . A nuisance variable
. A nuisance variable![]() , does not related to
, does not related to![]() .
. ![]() relate to
relate to ![]() at different levels. In terms of gene analysis, we can think of
at different levels. In terms of gene analysis, we can think of![]() ,
, ![]() and
and ![]() as underlying pathways, some of which are related to the disease measured by
as underlying pathways, some of which are related to the disease measured by![]() . We observed the gene expression levels,
. We observed the gene expression levels, ![]() , and would like to identify the related pathways.
, and would like to identify the related pathways.
We used all four Clustering Lasso methods. In all examples, the results from the four Clustering Lasso methods are close to each other. The prediction results from Lasso, CL2, CL4, Elastic Net, Ridge, and Weighted Fusion are summarized in Table 3 and Figure 3. In Figure 3, relative MSE was defined as the MSE of the corresponding method divided by the minimum MSEs from all the methods. We see that Clustering Lasso always performs better than the Lasso method, and it is close to or better than Ridge, Weighted Fusion and Elastic Nets, even under collinearity and group effect situations.
Table 4 shows the results of variable selection. The two numbers in each cell are the proportion of times an important factor is chosen and the proportion of times a false factor is chosen, respectively. We see that compared with Elastic Net, Weighted Fusion and Lasso, Clustering Lasso is superior at selecting important factors. However, like Weighted Fusion, it is more likely to over select variables than Elastic Net. In Example 2,
![]() (a) (b)
(a) (b)![]() (c) (d)
(c) (d)
Figure 3. Comparing the simulation results from the four examples. (a)-(d): Example 1-4.
![]()
Table 3. Mean (standard deviation) of MSE for the simulated examples based on the 100 iterations.
![]()
Table 4. Variable selection results for the simulated examples based on the 100 iterations. In each cell, the first number is the proportion of times a true factor is chosen and the second number is the proportion of times a false factor is chosen.
since all variables are highly correlated, Clustering Lasso cannot identify the most important variables. In comparison, Clustering Lasso performs very well in Examples 3 and 4, when clusters of variables play an important role in real model.
Finally, to show how Clustering Lasso chooses covariates in groups and the behavior of the coefficients for the selected variables, we illustrate the differences between Lasso and Clustering Lasso by a modified example from [1] . Let![]() ,
, ![]() and
and ![]() be three independent variables with the uniform
be three independent variables with the uniform ![]() distribution. The
distribution. The
response variable is generated as![]() . With the random error terms
. With the random error terms![]() , the nine
, the nine
observed predictors are
![]()
The variables![]() ,
, ![]() and
and ![]() are from group 1, with the direct effect
are from group 1, with the direct effect![]() .
.![]() ,
, ![]() and
and ![]() are from group 2, with the direct effect
are from group 2, with the direct effect![]() . The effect from
. The effect from ![]() on
on ![]() is much smaller than from
is much smaller than from![]() ―the coefficient for
―the coefficient for ![]() is 1 compared with 0.2 for
is 1 compared with 0.2 for![]() . Variables
. Variables![]() ,
, ![]() and
and ![]() are from
are from![]() , which does not relate to the response variable. The within-group correlations are almost 1, while the between group correlations are almost 0. Figure 4 shows the solution paths for Lasso, Elastic Net and CL2.
, which does not relate to the response variable. The within-group correlations are almost 1, while the between group correlations are almost 0. Figure 4 shows the solution paths for Lasso, Elastic Net and CL2.
We also use this simulation to compare the sensitivity and specificity of the listed methods in finding significant covariates. The simulation is repeated 100 times. Table 5 summarizes the number of times that the coefficients of ![]() are not zero. We find that the proposed Clustering Lasso of all versions can uniformly identify the important covariates while is less likely to select non-significant covariates than Lasso, Elastic Net and Weighted Fusion.
are not zero. We find that the proposed Clustering Lasso of all versions can uniformly identify the important covariates while is less likely to select non-significant covariates than Lasso, Elastic Net and Weighted Fusion.
![]()
Figure 4. Comparing the solution paths from Lasso, Elastic Net and Clustering Lasso.
![]()
Table 5. Number of times the coefficients are not zero based on the 100 repetitions.
6. Conclusions and Future Works
We find that the Clustering Lasso, is a novel predictive model that produces sparse model with good predictive performance, while encouraging group effects. The empirical results from a two-class microarray data classifica- tion problem and several simulation studies on regression problems show that Clustering Lasso has very good predictive performance and is superior to the Lasso method.
The method was proposed to encourage group effects so that clustered variables are selected together in a model. Clustering Lasso can automatically select groups of variables. If the structural correlation matrix used for regularization is block diagonal matrix, Clustering Lasso is equivalent to the group Lasso proposed by [7] . However, if the relationships among the variables are complicated, we have to simplify the structural correlation matrix to obtain sparse models. We proposed some shrinkage steps to build the desired structural correlation matrix. Rotating the
eigen
vectors or adapting techniques such as sparse component analysis can also help for this purpose. As a next step, we will use the Clustering Lasso method in the spatial analysis, so that we can maintain the important spatial correlations while selecting sparse models.
Acknowledgements
We thank
Mrs.
Patricia
Andrews
for editing the paper.