Phylogeny of Bacteria from Steelmaking Wastes and Their Acidic Enrichment Cultures ()
1. Introduction
Steelmaking activities have been particularly intensive during the 20th and 21st centuries, resulting in the generation of huge amounts of waste (approximately 700 kg of waste per ton of steel produced) including the presence of metals such Zn, Cu, and Cr [1] . Indeed, most of the waste is being left without proper management all over the world and without any management whatsoever in Brazil and perhaps in other countries as well. The release of these wastes to the environment leads to contamination and consequent human exposure to the metals present in these rejects. The World Steel Association reported an increase of 6.8% in steel production in 2011 resulting in 1.527 billion tons, of which Brazil contributed about 35.2 million tons (www.iabr.com.br). Thus the waste generated by this industry can no longer be ignored. First steps towards solutions for its management would be the search for suitable technologies to enable the removal of metals from these wastes.
Bioleaching of metals has gained increased attention since it is innovative, environmentally friendly, and economical [2] . Indeed, bioleaching has been considered as an alternative strategy for the extraction of metals from complex ores or wastes, which may reduce costs up to 80% when compared with traditional chemical techniques [3] . This biotechnological process is based on the ability of microorganisms to oxidize ferrous iron and/or reduce sulfur compounds [4] . The predominant metal-sulfide-dissolving bacteria that have been extensively used for the bioleaching of sulfide minerals with commercial interest are extremely acidophilic [5] [6] although heterotrophic bacteria can also contribute to metal leaching [7] .
The process of production of steel involves many stages, from reduction of iron ore at temperatures reaching 2400˚C (in a blast furnace) to metal plating, which uses a large amount of Zn to protect the steel from corrosion. Overall, the wastes contain tramp metals such as Zn, Cu, and Cr preventing their recycling for steel production due to possible damage to industry furnaces by these metals. Nevertheless, Zn is economically important due to its anti-corrosive and pharmaceutical properties. In this study, we identified and investigated the ability of heterotrophic bacteria from steelmaking wastes to survive in acidic conditions and to promote leaching of zinc.
2. Materials and Methods
2.1. Sampling and Chemical Composition of the Wastes
Steelmaking waste samples were collected at Usiminas (Ipatinga, Minas Gerais state, Brazil) using sterilized bottles. The wastes studied were crude thin sludge waste (TS), constituted by fine solid particles resulting from the Linz-Donawitz converter (or BOF), and crude sludge from treatment of electroplating effluent waste (STEE), both of which were released into the environment three days before sampling. X-ray fluorescence spectrometer (PW 2510 Sample Changer, Philips) analysis of the wastes revealed 1.9% and 29.8% Zn, 57% and 5.5% Fe, and 11% and 40% Ca in the TS and STEE wastes, respectively. The efficiency in Zn extraction was compared using an unpaired t-test, performed by PAST data analysis package. The level of significance was considered at p ≤ 0.05.
2.2. Acidic Enrichment Culture
Acidic enrichment cultures were established by blending 10 g of the separate TS and STEE wastes into 100 mL of Leathen medium [8] , and were respectively named TSC and STEEC. Prior to the bioleaching experiments the pH values of the crude STEE and TS wastes were 7 and 5, respectively. To prepare primary enrichment of heterotrophic bacteria, crude TS and STEE wastes were added to flasks and then treated with H2SO4 to reduce the pH value to 2 and thereby provide adequately acidic conditions for bacterial growth. The flasks were then incubated at 30˚C with agitation (200 rpm) for 40 days. The pH was monitored daily and adjusted with H2SO4 as needed. At the end of this period, we obtained the primary bacterial enrichment samples from each type of waste, TSC and STEEC.
2.3. Bioleaching Assay
Bioleaching assays were performed in 500 mL Erlenmeyer flasks. Each flask contained 180 mL of Leathen medium, 10 g of autoclaved waste (5% w/v), and 20 mL of TSC or STEEC (10% v/v). The flasks were incubated at 30˚C with agitation (200 rpm) for 30 days. The pH value in the leaching solution was kept constant (pH 2) throughout the leaching process by adding H2SO4 as need. After this period, the wastes were filtered, dried at 110˚C for one hour, and the concentration of Zn was measured by X-ray fluorescence spectrometer. These samples were henceforth called TSB and STEEB depending on the origin of inoculum used to start the bioleaching assay.
Non-inoculated controls consisted of autoclaved crude TS and STEE acidified and subjected to the same conditions as the TSC and STEEC tests. However, the pH of control flasks was monitored daily and continuously adjusted to pH 2 with H2SO4.
2.4. DNA Extraction and PCR Amplification of 16S rRNA Gene
Total DNA from waste (crude TS and STEE) and primary enrichment (STEEC and TSC) samples were isolated by using a MaxTM Power DNA Isolation Kit for soil and water (MO Bio Laboratories) according to the manufacturer’s instructions. The DNA samples were stored at −20˚C until further processing.
The bacterial 16S rRNA gene fragment was amplified using touchdown PCR according to Freitas et al. [1] , using the primer set 8f (5’AGAGTTTGATCMTGGCTCAG 3’) and 907r (5’ACGGHTACCTTGTTACGACTT 3’) [9] .
2.5. Cloning, Sequencing, and Clone Library Analysis
Bacterial 16S rRNA gene fragments were gel-purified using the Silica Bead DNA Gel Extraction Kit (Fermentas, Canada), cloned into the vector pJET1.2/blunt (Fermentas, Canada) according to the manufacturer’s instructions, and transformed into electrocompetent Escherichia coli XL1Blue. Partial 16S rRNA gene sequences were obtained using the pJET1.2 forward and reverse primers and a Mega BACE 1000 capillary sequencer (GE Healthcare, United Kingdom) according to the manufacturer’s instructions. Further, the sequences were checked for quality, aligned, and edited to produce a consensus using the programs Phred v. 0.020425 [10] , Phrap v. 0.990319 [11] , and Consed 12.0 [12] . Chimeras were checked and omitted using Bellerophon software (http://comp-bio.anu.edu.au/bellerophon/bellerophon.pl). Closely related sequences from Greengenes [13] were identified by blast tool using the Silva database. Phylogenetic relationships were inferred with ARB software [14] . The operational taxonomic units (OTUs) were set at 97% level identity using the DOTUR software [15] . Library coverage was calculated using the equation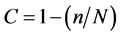 , where n denotes the number of unique OTUs and N is the number of sequences analyzed in the library [16] . The diversity of the OTUs was further examined using DOTUR software, LIBSHUFF statistics [17] , and rarefaction analysis. A comparative analysis was performed in order to detect OTU sequences shared among the four libraries. This analysis was performed using the DOTUR software [15] to detect sequence similarity at 97% level. The partial 16S rRNA gene sequences generated were deposited in the GenBank database under accession numbers KC164772-KC164863.
, where n denotes the number of unique OTUs and N is the number of sequences analyzed in the library [16] . The diversity of the OTUs was further examined using DOTUR software, LIBSHUFF statistics [17] , and rarefaction analysis. A comparative analysis was performed in order to detect OTU sequences shared among the four libraries. This analysis was performed using the DOTUR software [15] to detect sequence similarity at 97% level. The partial 16S rRNA gene sequences generated were deposited in the GenBank database under accession numbers KC164772-KC164863.
3. Results
3.1. Bioleaching of Zinc by Bacteria from Steelmaking Waste
In an attempt to extract Zn from the steelmaking wastes (STEE and TS) bioleaching assays were performed using bacteria from enrichment cultures of these wastes (STEEC and TSC). During bioleaching experiments the pH initially increased over the first seven days, being thus adjusted to pH 2 as indicated in the Materials and Methods section. After this time, acidity remained stable in the test flasks but not in the control flasks, which needed pH adjustments throughout the study period. Bioleaching efficiency was calculated by difference between the Zn contents in the crude TS and STEE samples and TSB and STEEB residues determined by X-ray fluorescence analysis. The data obtained revealed that STEEB (76%) and TSB (53%) presented efficiency in Zn extraction. Chemical leaching of Zn in the control flasks reached 82% and 47% for the STEE and TS wastes, respectively. Both leaching assays were equally efficient to remove Zn (p = 0.02).
3.2. Phylogenetic Affiliation
To reveal the phylogenetic identity of the bacteria, 16S rRNA gene clone libraries were constructed from crude TS and STEE wastes and from the TSC and STEEC enrichments. A total of 324 partial 16S rRNA gene sequences were obtained upon removal of chimeric sequences. These sequences were clustered into 94 OTUs spanning six bacterial phyla, mostly represented by cultivated heterotrophic bacteria. Clone libraries coverage accounted for >67% of the bacterial diversity. Rarefaction curves generated from our data did not reach an asymptote, indicating an amount of undetected diversity, especially for the STEEC (Figure 1). The phylogenetic distributions of the OTUs and the resulting phylogenetic trees are shown in Figure 2 and online Resources 1-4, respectively.
![]()
Figure 1. Rarefaction analysis of bacterial 16S rRNA gene sequences from crude sludge from treatment of electroplating effluent (STEE), enrichment culture from treatment of electroplating effluent (STEEC), cru- de thin sludge (TS), and enrichment culture from thin sludge (TSC). The total number of sequenced clones is plotted against the number of OTUs observed in the same library. The OTUs were defined at the ≥97% identity level (species level).
![]()
Figure 2. Phylogenetic ARB affiliation of bacterial 16S rRNA genes. The numbers indicate the percentage representative of each phylum in the library. (A) crude sludge from treatment of electroplating effluent (STEE); (B) enrichment culture from treatment of electroplating effluent (STEEC); (C) crude thin sludge (TS); and (D) enrichment culture from thin sludge (TSC).
![]()
Resource 1.Phylogenetic neighbor-joining tree of bacterial OTUs from crude sludge from treatment of electroplating effluent (STEE) constructed using the ARB software.
![]()
Resource 2.Phylogenetic neighbor-joining tree of bacterial OTUs from enrichment culture from treatment of electroplating effluent (STEEC) constructed using the ARB software.
![]()
Resource 3.Phylogenetic neighbor-joining tree of bacterial OTUs from crude thin sludge (TS) constructed using the ARB software.
![]()
Resource 4.Phylogenetic neighbor-joining tree of bacterial OTUs from enrichment culture from thin sludge (TSC) constructed using the ARB software.
The Proteobacteria and Firmicutes phyla contained most of the OTUs identified in the crude STEE clone library (Figure 2(A)). Proteobacteria was represented by the Gammaproteobacteria (50%), Betaproteobacteria (37%), and Alphaproteobacteria (13%) classes.
Clone library analysis from the STEEC revealed that the acidic pH promoted the emergence of Deinococcus-Thermus and disappearance of Actinobacteria (Figure 2(A) and Figure 2(B)). Overall, the 27 OTUs harbored two phyla in common with the STEE clone library: Proteobacteria and Firmicutes, with a strong dominance of Proteobacteria, represented by Gammaproteobacteria (54%), Betaproteobateria (38%), and Alphaproteobacteria (8%) classes.
According to the phylogenetic analysis of the 16S rRNA gene sequences from the crude TS clone library, 30 OTUs were affiliated with the Proteobacteria, Firmicutes, Actinobacteria, and Cyanobacteria phyla (Figure 2(C)). Proteobacteria was the most abundant phylum, Firmicutes and Actinobacteria contributed evenly to bacterial community, whereas Cyanobacteria was present in lower ratios. Gammaproteobacteria (40%), Alphaproteobacteria (33%), and Betaproteobacteria (27%) classes were found.
The TSC clone library was composed of OTUs affiliated with Proteobacteria, Firmicutes, and Actinobacteria, with extensive variation in the proportional distributions of these phyla (Figure 2(D)). Gammaproteobacteria (70%), Alphaproteobacteria (15%), and Betaproteobacteria (15%) classes were also present. Tables 1-4 show the classification of OTUs from all libraries down to genus and species level.
3.3. Comparisons of Bacterial Compositions Based on OTU Clustering
To determine the significance of differences between the clone libraries based on 16S rRNA gene sequences, we applied LIBSHUFF statistics, and the results revealed no significant differences in composition of bacterial communities.
To cluster sequences into OTUs and to distinguish between the shared and sample-specific OTUs, all sequence data were pooled together and analyzed using DOTUR (at >97% similarity). The OTUs were divided into three categories as plotted in a Venn diagram (Figure 3): core OTUs shared by all crude and enriched samples, OTUs shared by two or three samples, and sample-specific OTUs. Four bacterial communities shared 11 OTUs comprising the Escherichia and Chromobacterium genera as shown in the diagram. The genus Ochrobactrum was present only in the enriched samples of both waste types (STEEC and TSC). The diagram also reveals that all crude and enriched samples shared four bacterial communities (Chromobacterium, Escherichia, Bacillus, and Ochrobactrum).
The potential role of these bacterial communities in the Zn extraction processes from steelmaking wastes will be discussed in the following section.
4. Discussion
Environmental metal pollution is a serious problem and the treatment or recovery of desired metals from wastes is a major challenge for the sustainable use of non-renewable natural resources such as Zn. In this study, we performed assays under extremely acidic conditions, and showed that heterotrophic bacteria from steelmaking wastes were able to survive and had similar efficiency to extract Zn as in chemical leaching. However, the bi- oleaching assay could be considered more advantage since in this condition there was not need to addition acid
![]()
Table 1. Phylogenetic affiliation and distribution of bacterial 16S rRNA gene sequences analyzed from the STEE library.
![]()
Table 2. Phylogenetic affiliation and distribution of bacterial 16S rRNA gene sequences analyzed from the STEEC library.
to maintain the pH 2. The bioleaching process is often based on acidophilic bacteria such as Thiobacillus ferro- oxidans, Leptospirillum ferrooxidans, and Acidithiobacillus thiooxidans, which have been implicated as being the most applicable bacteria involved in operation of biological metal-removal processes including Zn [18] -[20] .
![]()
Table 3. Phylogenetic affiliation and distribution of bacterial 16S rRNA gene sequences analyzed from the TS library.
In contrast to these reports we reveal herein heterotrophic bacteria phylogenetically distinct from acidophilic bacteria.
It is well-established that metals can be removed from a variety of wastes by lowering the pH either by the
![]()
Table 4. Phylogenetic affiliation and distribution of bacterial 16S rRNA gene sequences analyzed from the TSC library.
![]()
Figure 3. Comparative analysis of the OTU sequences shared among the four libraries.
addition of acids or by the production of acids by bacterial activity. As expected, we found that a chemical Zn leaching process also occurred in our un-inoculated steelmaking waste samples.
The pH increase observed at the first seven days in both wastes, could be explained by consumption of H+ protons, which could be illustrated by the equation Fe2+ + 0.5O2 + 2H+ → Fe3+ + H2O [21] . Even during this period, bacterial growth could occur through the energy obtained in the Fe2+ oxidation. The Fe3+ formed would be further used as in the equation 4Fe3+ + ZnS + 2H2O + O2 → Zn2+ + 4Fe2+ + 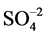 + 4H+, resulting in the release of protons and completing the iron redox cycling. Therefore, the combination of chemical reactions in acidic conditions, along with the emergence of bacterial metabolic activity favored the maintenance of a pH value of 2, at one week of leaching of the steelmaking samples analyzed. Thus, pH decrease in the bioleaching assays may be an indication of the bacterial activity enhancing the iron redox cycling and leading to an effective solubilization of Zn as suggested by Marhual et al. [22] .
+ 4H+, resulting in the release of protons and completing the iron redox cycling. Therefore, the combination of chemical reactions in acidic conditions, along with the emergence of bacterial metabolic activity favored the maintenance of a pH value of 2, at one week of leaching of the steelmaking samples analyzed. Thus, pH decrease in the bioleaching assays may be an indication of the bacterial activity enhancing the iron redox cycling and leading to an effective solubilization of Zn as suggested by Marhual et al. [22] .
Using culture-independent molecular and enrichment culture approaches, this study provided insight into the bacteria from STEE, TS, STEEC, and TSC by revealing their phylogenetic identity. Overall, the steelmaking wastes harbored few lineages, suggesting an unfavorable environment for the bacterial communities present in them. Proteobacteria was systematically the phylum predominant (from 50% to 87%) in all the clone libraries. According to Chen et al. [23] , in a metal-rich environment the proteobacterial account for up to 70% of 16S rRNA gene clone library sequences. Although the number of genera found was only eight, this ubiquitous phylum harbors several members associated with iron redox cycling and ability to leach metal in steelmaking wastes. Other phyla also disclosed herein have already been shown to harbor members with similar abilities [24] -[26] .
Diverse bacterial taxa can be present in a metal-rich environment. However, metal-tolerant bacteria appear to be present as primary heterotrophic colonizers of exposed minerals [27] . Moreover, Johnson and Roberto [28] reported that heterotrophic bacteria can obtain carbon sources through the wastes products produced by the autotrophs and play a role in the mineral degradation processes. Given these data, it is likely that Zn leaching bacteria found in our steelmaking waste sample are also heterotrophic.
The Chromobacterium, Escherichia, Bacillus, and Ochrobactrum genera were common in the primary enrichment of both clone libraries, suggesting that they could play a role in Zn bioleaching of steelmaking wastes. Previous studies reported bioleaching ability of Chromobacterium violaceum to mobilize diverse metals, including Zn, from solid and electronic materials [29] [30] . An unexpected result was the occurrence of a strong dominance of Escherichia in the TSC waste. Indeed, it is not clear why and how this dominance occurs in such a poor carbon source environment and therefore further studies will be needed to clarify this matter. One possibility is the versatile behavior of E. coli which is able to tolerate and rapidly adapt to diverse stress environmental conditions such as low pH and strongly carbon-limited, among others (reviewed by [31] ). Although Lin et al. [32] reported the ability of E. coli to survive at low pH, it is the first time that this genus has been connected with Zn extraction and the first time it has been reported in steelmaking wastes.
Representatives of Bacillus have been reported to efficiently solubilize metals and thus contribute to metal leaching without any benefit to themselves [7] [33] . Only one other study found Bacillus from steelmaking waste sources. That work, which was from our laboratory, reported an interesting dominance of this genus in Blast Furnace Sludge wastes [34] . We also found members of Ochrobactrum in our steelmaking wastes. Ozdemir et al. [35] reported that members of this genus are involved in the solubilization of other metals such as chromium, cadmium, and copper. However, we did not find such metals in our samples, indicating the ability of Ochrobactrum to solubilize metals other than those.
Aeromonas was only found in the STEEC clone library, where it shared dominance with Chromobacterium. Indeed, a previous study showed the presence of Aeromonas in uranium deposits [36] while another study revealed that members of this genus were able to reduce Fe3+, nitrate, and sulfate [37] .
This study is the first report of survival of heterotrophic bacteria from steelmaking wastes under extremely acidic conditions, such as Chromobacterium, Aeromonas, Escherichia, Bacillus, and Ochrobactrum. The data presented herein may be relevant for the management of this waste, particularly in the case of Zn extraction, making these bacteria candidates for future studies about metal bioleaching.
Acknowledgements
We thank FAPEMIG (Fundação de Amparo à Pesquisa do Estado de Minas Gerais), CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior), and CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico) for providing financial support. We also thank Usiminas, which provided the samples and the chemical characterization.
NOTES
*Corresponding author.