Aptamer-Based Extraction of Ergot Alkaloids from Ergot Contaminated Rye Feed ()
1. Introduction
Ergot alkaloids are biologically active compounds mainly produced by fungi of the genus Claviceps [1] [2] , which can grow on more than 400 plants and grasses; mainly forage and leading cereal crops worldwide such as wheat or sorghum [1] [2] . Claviceps fungi produce ergot alkaloids as secondary metabolites, which are toxic to humans and animals. Ergot alkaloids are based on a four-member ring, ergoline, which is mainly responsible for their biological activity [3] . Ergot alkaloids are undesirable in food and feed, as they can lead to a poisoning known as ergotism [4] . However, a few ergot alkaloids exhibit interesting medicinal properties [5] , and are produced on purpose by fungal contamination of cereal crops or produced by saprophytic cultures for instance [6] [7] . Chemical extraction techniques were developed in order to isolate ergot alkaloids [8] -[10] , and to quantify them [11] -[15] . However, these techniques are destructive for the samples and cannot be applied for the decontamination of ergot alkaloids from food and feed stuff. Therefore, other techniques were recently elaborated for the removal of ergot alkaloids from food and feed; such as molecularly imprinted polymers (MIPs) [16] [17] . Also, biological recognition elements were developed for ergot alkaloids, such as antibodies [18] [19] and aptamers [20] . These different recognition elements can be immobilized onto solid supports in order to obtain specific solid phase extraction systems. Different types of solid supports can be used, such as organic polymers or inorganic oxides for instance. Silica gel is interesting for this type of applications because it presents a high thermal and chemical stability and enables a wide variety of chemical modifications through the silanol groups dispersed on its surface [21] . The covalent coupling of organic molecules and biomolecules can be done by chemical pretreatments of silica particles with organosilanes, such as dichlorodimethylsilane (DDS) [22] [23] , 3-chloropropyltrimethoxysilane (CTS) [24] [25] , or 3-aminopropyltriethoxysilane (APTS) [26] . In the present study, we report the functionalization of silica particles with two different aptamers specific to ergot alkaloids, in order to construct a specific solid phase extraction system. The aptamer-functionalized silica gel was tested with a raw extract of contaminated rye flour. The presence of ergot alkaloids eluted from the aptamer functionalized silica gel was determined by quadrupole-time-of-flight mass spectrometry (LC-MS-QTOF) analysis. It was observed that the aptamer-functionalized silica gel could successfully extract three main ergot alkaloids from the sample; namely ergosine, ergokryptine and ergocornine. The use of DNA-grafted silica gels has already been reported in sensing assays [26] -[29] and in extraction systems [30] [31] . However, the realization of aptamer-functionalized silica gel for the specific extraction of ergot alkaloids is reported for the first time in the present article.
2. Materials and Methods
2.1. Choice of Single-Stranded DNA Aptamers
An aptamer, named aptamer M3.2, having a dissociation constant of 44 nmol2/L2, was previously selected for the ergoline group and was reported in our previous research article [20] . The original aptamer M3.2 consists of 80 bases: but in this experiment, only a fragment of 52 bases containing the predicted binding sites was used for the functionalization of silica gel, taking care of keeping the same conformation of the binding sites. The following 3'-aminated version of the shortened aptamer M3.2 was used 5'-GGTCAGATGTCCGTCAGCCCCGATCGCCATCCAGGGACTCCCCCCTACTGCC-3'-NH2. Another aptamer selected for ergot alkaloids, named L5.5 and having a dissociation constant of 660 nmol/L, was also tested in this study. The 3'-aminated aptamer L5.5 was used, having the following sequence 5'-AGCAGCACAGAGGTCAGATGGGCAGGATACAACACGTTACTATCCACTCTGCACCGGCGGCCTATGCGTGCTACCGTGAA-3'-NH2. The secondary structures of aptamers M3.2 and L5.5, determined by using IDT Oligoanalyzer software [32] , are shown in Figure 1.
2.2. Silica Functionalization with Aminated DNA
A narrow particle range silica gel of 10 - 20 µm (Merck, Germany) was used to prepare the aptamer-functionalized silica gel. For each sample, 300 mg of silica gel were placed in Eppendorf tubes.
Sample 1 was kept as unmodified silica gel in order to be used as a reference material. Sample 1 was washed two times with 500 µL of ultrapure MQ water and placed in 500 µL of MQ water.
The silica gels of Samples 2 and 3 were activated by a treatment with DDS, in order to enable functionalization with aminated molecules [22] . The chemical functionalization of DDS activated silica gel with aminated DNA is represented in Figure 2. The silica gels of Samples 2 and 3 were first washed two times with 200 µL of diethyl ether and then placed in 500 µL of 5% diethyl ether solution of DDS (Sigma-Aldrich, Belgium), and the solutions were gently stirred for 30 min at room temperature (RT). Then, the solutions were poured in centrifugation columns and the solutions were centrifuged at 13,000 rpm during 1 min. The silica gels were washed two times with 300 µL of methanol and centrifuged in centrifugation columns. Finally, the silica gels were washed three times with 300 µL of MQ water and placed in 300 µL of MQ water.
For the preparation of Sample 2, 0.63 mg of shortened aptamer M3.2 (40 nmol) (Eurogentec, Belgium) was placed in 200 µL of MQ water. Sample 3 was prepared by placing 1.6 mg of 3'-aminated aptamer L5.5 (65 nmol) (Eurogentec, Belgium) in 200 µL of MQ water. The two solutions of 3'-NH2 modified aptamers were mixed with the solutions of DDS treated silica, and the mixtures were gently stirred for 24 h at RT. The solutions were then centrifuged at 13,000 rpm during 2 min. The recovered silica gels were washed three times with 500 µL of MQ water and finally placed in 500 µL of MQ water.
2.3. Sample Preparation
A sample of ergot contaminated rye feed having a known concentration of ergot alkaloids was used for the extraction of ergot alkaloids. A sample of 0.5 g of the ergot contaminated flour sample was placed in 5 mL of 0.1


Figure 1. Secondary structures of shortened version of aptamer M3.2 (left) and aptamer L5.5 (right).
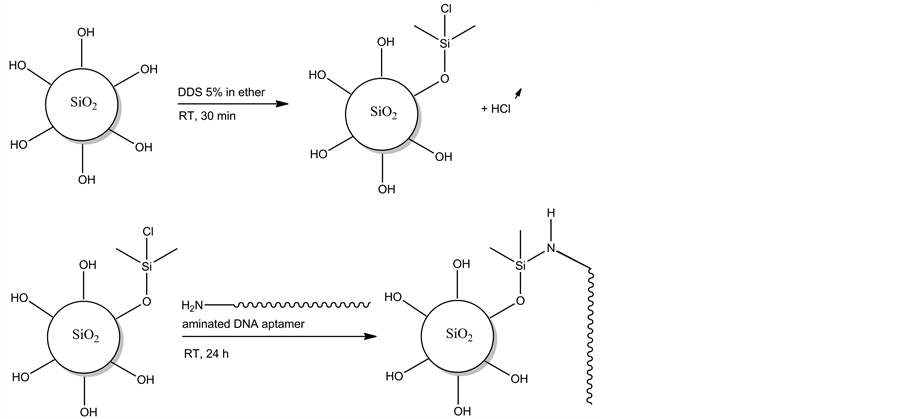
Figure 2. Chemical representation of silica gel functionalization with aminated DNA using DDS as silane-coupling agent.
M HCl and gently stirred for 1 h at RT. Then, the solution was transferred to Eppendorf tubes, which were centrifuged at 13,000 rpm for 2 min at RT. The supernatant was taken and placed in clean vials.
2.4. Specific Extraction of Ergot Alkaloids Using Aptamer-Functionalized Silica Gels
For each silica sample, 500 µL of rye feed extract were used. The reference sample was prepared by adding 500 µL of rye feed extract to the solution of unmodified silica gel. Sample 2 was prepared by mixing 500 µL of rye feed extract with the solution of aptamer M3.2 functionalized silica gel. Finally, Sample 3 was prepared by mixing 500 µL of rye feed extract with the solution of aptamer L5.5 functionalized silica gel. The three solutions were gently mixed for 1 h at RT. After centrifugation in centrifugation tubes, the supernatants were discarded, and the silica gels were washed three times with 500 µL of MQ water. For the elution step, the silica gels were recovered and placed in clean Eppendorf tubes with 500 µL 0.1 M HCl, and were then gently stirred at 70˚C for 15 min. The silica gels were centrifuged in centrifugation tubes at 13,000 rpm for 1 min and the supernatants were kept for the LC-QTOF-MS analysis.
2.5. LC-QTOF-MS Analysis
Ergot alkaloids were separated and identified by LC-QTOF-MS having a 1290 series High Pressure Liquid Chromatography (HPLC) system coupled to a 6530 quadrupole-time-of-flight mass spectrometer (QTOF-MS) (Agilent Technologies). Chromatographic separation was achieved using a Zorbax Eclipse Plus C18 column (100 mm ´ 2.1 mm ´ 1.8 µm, Agilent Technologies) and the following mobile phase composition: water 0.1% formic acid in water (A) and 0.1% formic acid in acetonitrile (B). Gradient elution was as follows: initial 5% B, linear change of B to 99%, from 0 to 5.0 min followed by isocratic elution at 99% B from 5.0 to 8.0 min, then linear change of B% to 5% until 8.1 min, then maintained at 5% B until 12.0 min in order to re-equilibrate the column. The flow rate was 0.3 mL/min and the injection volume 5 µL. Total run time was 12.0 min. The column was kept at a constant temperature of 45˚C. The QTOF-MS was run in ESI positive mode scanning m/z from 50 to 1000 amu at a scan rate of 3 spectra/s. An Agilent Jetstream source was used with the following parameters: gas temperature 275˚C, gas flow 8 L/min, nebulizer pressure 40 psi, sheath gas temperature 325˚C, sheath gas flow 11 L/min. Nozzle, capillary, fragmentor, and skimmer voltages were set to 0 V, 3500 V, 110 V, and 65 V, respectively. The instrument was calibrated during run times by monitoring positive ions with m/z 121.0508 and 922.0098. The Mass Hunter (Agilent Technologies) software was used for data acquisition and processing.
3. Results and Discussion
The ergot alkaloid content of the sample of ergot contaminated rye feed given by the LC-MS analysis performed by Diana Di Mavungu et al. [33] is shown in Table1 The total ergot alkaloid concentration of this sample given by this analysis was 1081 µg/kg, representing a moderately high contaminated ergot sample. Ergotamine, ergosine, ergocornine and ergokryptine were the main ergot alkaloids of this sample according to this study.
The ergot alkaloids eluted from the silica gels were analyzed by LC-QTOF-MS analysis. LC-QTOF-MS technique was used because it provides a high mass accuracy (typically 2 to 10 ppm error) and a good resolution (>20,000), which is necessary to generate a formula for an unknown compound with a given m/z value. Another advantage of the LC-QTOF-MS is its ability to fragment compounds, which provides high resolution mass spectra of the product ions allowing the confirmation of the generated formula. The screening of the six main ergot alkaloids was based on a self-made comma separated value (csv) database. The accurate masses given with four digits were compared to the theoretical monoisotopic masses, with the experimental error given in ppm.
In the LC-QTOF-MS analysis of the samples after extraction using the aptamer-functionalized silica gels, the following data were analyzed: the presence of the six main ergot alkaloids, the presence of the ergot alkaloid precursor fragment at m/z 223.1283, and finally the presence of other compounds than ergot alkaloids. The samples were analyzed in positive ion mode; and the hydrogen atom mass was subtracted in the final results. Table 2 shows the compounds found in the three samples tested. In Sample 1, resulting from the elution from non-functionalized silica gel, no ergot alkaloid could be found, neither the ergot alkaloid precursor fragment. No other compound was detected in this sample. In Sample 2, resulting from the elution from aptamer M3.2 functionalized silica gel, two ergot alkaloids, were found: ergosine at m/z 547.2874 (theoretical m/z 547.2794) and ergokryptine 575.3085 (theoretical m/z 575.3107). The fragment at m/z 223.1283 was found for both the ergot

Table 1 . LC-MS analysis of the ergot alkaloid content of the ergot contaminated rye sample [12] [33] .

Table 2. LC-QTOF-MS analysis of the ergot alkaloid content after elution from silica gels.
alkaloids detected. No other compound or impurity were found, or were present at a very low level not detectable by this analysis. In Sample 3, resulting from the elution from aptamer L5.5 functionalized silica gel, three ergot alkaloids were found: ergosine at m/z 547.2797 (theoretical m/z 547.2794), ergokryptine at m/z at 575.3112 (theoretical m/z 575.3107) and ergocornine at m/z 561.2916 (theoretical m/z 561.2951). The ergot alkaloid precursor fragment at m/z 223.1283 was found for the three ergot alkaloids detected in the sample. No other compound was detected in this sample as well. The experiment was repeated three times and gave similar results. These results show that the presence of aptamers in the silica gel allowed the specific extraction of ergot alkaloids from the sample. However, the other ergot alkaloids present in the sample; such as ergotamine, ergometrine and ergocristine; were not retained by the aptamer-functionalized silica gels, or were present at a very low level not detectable by this analysis.
In this study, it was observed that aptamers selected for ergot alkaloids were highly specific, having a molecular recognition restricted to a few related chemical compounds within the ergot family group. In comparison to other biorecognition elements developed for ergot alkaloids, similar specificity was obtained with antibodies [34] . Having a similar molecular affinity towards ergot alkaloids, aptamers present several advantages over antibodies, such as their chemical robustness and their animal-friendly synthetic production. In comparison to MIPs, it was observed that aptamers were more specific and had a more restricted range of recognition [16] [17] . Depending on the use of the extraction system, it can be said that MIPs are suited for a general extraction of ergot alkaloids, while aptamers can be used for a more specific extraction of certain ergot alkaloids. It is also possible to envisage a more complete extraction system by mixing different aptamer-functionalized silica gels. This would allow the modulation of the extraction process to a more or less important range of molecules within the ergot alkaloid family. Concerning the extraction solution, only 0.1 M HCl was used; which can be easily neutralized by a 0.1 M basic solution. This study shows the applicability of aptamers for extraction purposes, offering a new range of possibilities for the specific extraction of toxins, natural compounds or pollutants from food or environmental matrices.
4. Conclusion
Aptamer-functionalized silica gels were realized in order to extract ergot alkaloids from ergot contaminated rye feed. The ergot alkaloids eluted from the aptamer-functionalized silica gels were measured by LC-QTOF-MS. It was shown that the aptamer-functionalized silica gels could successfully be used to extract three ergot alkaloids from the sample, namely ergosine, ergokryptine and ergocornine. The other compounds present in the sample were not retained by such a system. The DNA aptamers were neither denatured by the silica functionalization, nor by the acidic solution used for the extraction of ergot alkaloids. Although aptamers were mainly developed for sensing purposes, this study shows that it is also possible to use aptamers for the specific extraction of compounds. This type of system can be easily applied for food clean-up, extraction of toxins or contaminants from various environmental or food samples, as well as the specific extraction of natural compounds from turbid matrices.
Acknowledgements
This study was funded by the Belgian Federal Public Service of Health, Food Chain Safety and Environment (FOD) project Ergot RF6204. The authors thank Dr. Abdelkrim Azmi for his help and support.
NOTES
*Corresponding author.