Influence of GSTT1, GSTM1 and GSTP1 Polymorphisms on the Development of Breast Cancer ()
1. Introduction
Cancer arises from the accumulation of genetic and epigenetic alterations; the most common alterations include amplification of proto-oncogenes and loss of chromosomal material; this causes loss of control of the cell cycle and sidestepping DNA repair and apoptosis, al-lowing the cell to replicate uncontrollably [1] [2] .
It is estimated that each year more than 10 million women worldwide are diagnosed with breast cancer and about 7 million deaths directly or indirectly occur due to this malignancy, which remains the leading cause of cancer death in women. There were 4206 deaths from breast cancer in Mexico in 2005; this means that about 12 Mexican women die from it every day. In year 2006, incidence of breast cancer outdid cervical cancer and ranked as the second leading cause of death among women 30 to 54 years old [3] -[5] .
Age, sex, immune status, preexisting conditions, the environment, nutrition and genetic load are some factors that influence susceptibility to cancer; genetic load being one of the most important ones for the interaction between etiological agents and subjects [3] [5] .
The presence of polymorphisms or variations in the base sequences of metabolic genes give rise to variable enzyme activity which leads to differing abilities in the metabolism of xenobiotics [6] , of which the glutathione S-transferase family (GST) is a part. It consists of several genes coding for a group of isozymes involved in Phase II metabolism, protecting the cell from oxidative injury [7] -[10] . The most studied GST subclasses in mammals are Mu (μ), Pi (π) and Theta (θ) [11] [12] .
The GSTM1 (μ) gene, located on chromosome 1p13.3, consists of 8 exons, with a length of 4.2 kb and 4 allelic variants: A (wild), B, C and 0 (zero). GSTM10 is the most frequent polymorphism, resulting in the loss of enzymatic activity [11] [13] -[15] .
The GSTT1 (θ) gene is located on chromosome 22p11.2; it consists of 6 exons and is flanked by two homologous regions HA3 and HA5. It has two allelic variants: wild GSTT1a and GSTT10, null. This polymorphism is due to a homologous recombination of the HA3 and HA5 regions resulting in a deletion of 5.4 kb [13] [15] -[17] .
GSTP1 gene (π) is located on chromosome 11q13; it consists of nine exons, with a length of 3.2 kb and has three allelic variants: wild allele GSTP1a, the polymorphic allele GSTP1b, located in exon 5 which is the result of the mutation of ATC (Ile) to GTC (Val) at codon 104. The third allele, GSTP1c, also polymorphic, has the same mutation at codon 104 as GSTP1b and, additionally, a second mutation in exon 6, consisting of a base substitution of GCG (Ala) to GTG (Val) at codon 113 [8] [15] [18] -[21] . The enzymes coded by these alleles have been implicated in the regulation of cell proliferation and apoptosis via direct interaction with c-Jun N-terminal kinase (JNK) [22] [23] .
Null or heterozygote genotypes of different classes of GSTs have been linked to susceptibility to developing lung, bladder, colon, skin, liver and breast cancer. The pharmacological interest of GSTs is their influence on the development of cancer on one hand, and on the response to therapy on the other [13] -[15] [19] [24] [25] . The aim of this study was to determine the frequency of M1, T1 and P1 polymorphisms in patients with breast cancer and to determine its association with the development of this malignancy.
2. Materials and Methods
2.1. Studied Group
This study was cross-sectional, descriptive and comparative in nature. The patient group consisted of 22 women with a clinical diagnosis of breast cancer at different stages of from the ISSEMyM State Oncology Center (Toluca, México). The control group was integrated by 30 healthy women in the same age range, native from the Toluca valley, Mexico. Participation was by invitation and voluntary; those who agreed to participate signed a letter of informed consent. A 3 mL sample of peripheral blood from each participant was drawn into a Vacutainer tube with heparin, which was kept refrigerated until processed.
2.2. DNA Extraction
DNA extraction was performed using the Quick-gDNAMiniPrep Kit (Zymo Research). The products were verified by horizontal electrophoresis in 1% agarose.
2.3. PCR for GSTT1 and GSTM1
Identification of polymorphisms of T1 and M1 was performed by multiplex PCR with CYP1A1 gene as control. The mixture had a final volume of 10 μL, containing 1 μL 5x PCR buffer (Promega), 0.6 μL of 25 mM MgCl2 (Promega), 1 μL 50 μM dNTP (Fermentas), 0.3 μL of molecular biology grade H2O, 0.1 μL 5 U/μLTaq Polymerase (Promega), 1 μL of DNA template and 1 μL of each primer CYP1A1-f, CYP1A1-r, GSTM1-f, GSTM1-r GSTT1-f, GSTT1-r, all primers had a concentration of 30 pm, Table 1. PCR conditions for these amplifications were: 5 minutes at 94˚C, denaturation 1 minute at 94˚C, alignment 1 minute at 59˚C, extension 1 minute at 72˚C and a final elongation of 5 minutes at 72˚C, for 35 cycles. PCR products were verified by horizontal electrophoresis using 1.5% agarose. The determination of the polymorphism of GSTT1 was based on the presence of a 480 bp band, corresponding to wild GSTTl, while its absence denoted a null GSTT1. Similarly, for GSTM1 the presence of a 215 bp band indicated wild GSTM1 while its absence implied null GSTM1. Finally, a 312 bp band, corresponding to CYP1A1, was always present and was employed as a PCR control gene [15] [25] -[27] .
2.4. PCR-RFLP for GSTP1
Identification of P1b (exon 5) and P1c (exon 6), was carried out in two separate PCRs, one for each polymorphism, both with a final volume of 25 μL in order to obtain a sufficient amount of product for subsequent enzymatic digestions. The mixture for the PCR reaction contained 2 μL of template DNA, 5 μL of 5x PCR buffer (Promega), 1.5 μL of 25 mM MgCl2 (Promega), 0.1 μL of 5 U/μLTaq Polymerase (Promega), 0.5 μL 50 μM dNTPs (Fermentas), 13.9 μL molecular biology grade H2O and 1 μL of each primer according to the gene to be amplified GSTP1b-f, GSTP1b-r GSTP1c-f or GSTP1c-r; all primers had a concentration of 30 pm, Table 1. For GSTP1c PCR, the conditions were the same as those used for GSTT1 and GSTM1, while conditions for GSTP1b PCR were: 5 minutes at 94˚C, denaturation 1 minute at 94˚C, alignment 1 minute at 62˚C, extension 1 minute at 72˚C and a final elongation of 5 minutes at 72˚C for a total of 35 cycles [28] .
For the identification of P1 polymorphisms, enzymatic digestions were carried out using BsmAI (Fermentas) for P1b (exon 5) and AciI (Fermentas) for P1c (exon 6). The digestion mixture consisted of 17 μL molecular biology grade H2O, 2 μL Fast-Digest Green buffer (Fermentas), 1 μL Fast-Digest Enzyme (Fermentas) and 10 μL PCR product DNA, for a total volume of 20 μL. Both digestion reactions were incubated at 37˚C for 5 minutes. The digestion products were verified by horizontal electrophoresis, using a 2% concentration of agarose. The identification of polymorphisms was based on the presence of DNA fragments of different sizes. For exon 5, 176 bp, 91 bp and 85 bp fragments correspond to heterozygote P1b while 91 bp and 85 bp P1b fragments correspond to homozygote and a 176 bp fragment corresponds to wild P1a [26] -[28] . Regarding exon 6 polymorphisms, the 332 bp fragment corresponds to homozygote P1c; three fragments, 332 bp, 174 bp and 158 bp correspond to heterozygote P1c and two fragments, 158 bp and 174 bp, to wild P1a [29] .

Table 1. Primers for GSTM1, GSTT1, CYP1A1 and GSTP1 genes.
2.5. Statistical Analysis
A X2 test was used to compare the frequencies of polymorphisms of GSTT1, GSTM1 and GSTP1 between the two groups, using the Sigma Stat 3.5 program.
3. Results
3.1. Socio-Demographic Attributes
The average age of patients with breast cancer was 49.4 years, ranging from 30 to 66. In the control group, the average age was 46 years, ranging from 30 to 65 years.
With respect to patients with breast cancer, the majority, 91.4%, had infiltrating ductal type and 8.6% were of the infiltrating lobular type.
3.2. Genotyping
Frequencies of polymorphisms obtained for the breast cancer group were: 77% wild GSTT1 and 33% null; 68% was wild GSTM1 and 32% null, while in the control group 60% was wild GSTT1 and 40% null; 37% was wild GSTM1 and 63% null. The statistical analysis showed a significant X2 difference in GSTM1 polymorphisms between the two groups. These results are presented in Figure 1.
The frequencies of GSTP1b polymorphisms (exon 5) in the breast cancer group were: 19% for wild P1a/P1a, 63% heterozygote P1a/P1b and 18% P1b/P1b homozygote; for GSTP1c (exon 6), 59% was wild P1a/P1a, 41% heterozygote P1a/P1c and 0% homozygote P1c/P1c. In the control group frequencies for GSTP1b (exon 5) were 27% wild P1a/P1a, 53% heterozygote P1a/P1b and 20% homozygote P1b/P1b; regarding GSTP1c (exon 6), 90% was wild P1a/P1a and 10% heterozygote P1a/P1c; homozygote P1c/P1c was not found. The X2 statistical analysis showed significant differences for heterozygote P1a/P1c and wild P1a/P1a GSTP1c polymorphisms, between the two groups.
Once genotype frequencies were individually determined, we proceeded to evaluate combinations of them. We found that for the breast cancer group, the most common genotypes were, firstly, wild GSTM1, wild GSTT1, homozygote (P1b/P1b) GSTP1 exon five and exon six wild GSTP1 (P1a/P1a) with 18% and secondly, with 15%, wild GSTM1, wild GSTT1, exon five heterozygote GSTP1 (P1a/P1b) and exon six heterozygote GSTP1 (P1a/P1c). While the two most common genotypes in the control group were wild GSTM1, wild GSTT1, exon five heterozygote GSTP1 (P1a/P1b) and exon six wild GSTP1 (P1a/P1a) with 20% and secondly, with 17%,
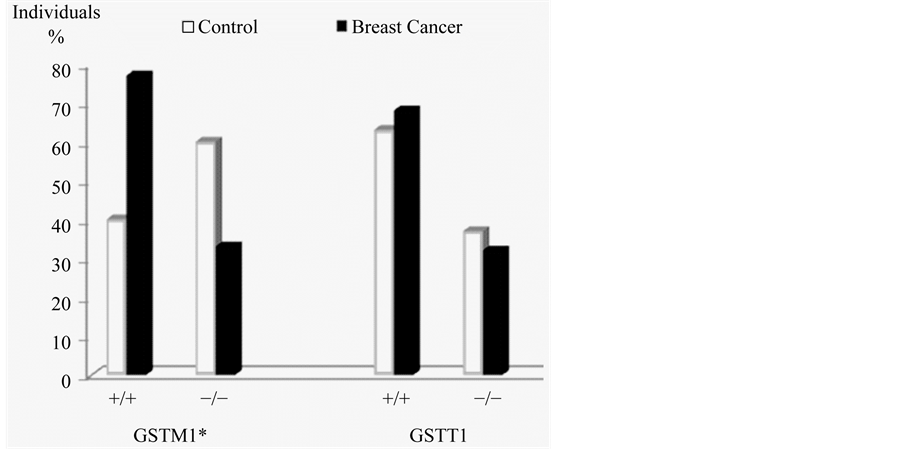
Figure 1. Frequencies of GSTM1 and GSTT1 polymorphisms. *Significant difference for GSTM1 between groups, p ≤ 0.05.
GSTM1 null, wild GSTT1, exon five heterozygote GSTP1 (P1a/P1b) and exon six wild GSTP1 (P1a/P1a). The details of these results are presented in Figure 2 and Table 2.
4. Discussion
Breast cancer is a disease of multifactorial etiology that is associated with age, for this reason the individuals in the control group for this study were within the age range of patients, to prevent age from being a misleading factor.
The presence of polymorphisms in susceptibility genes such as the GST family has been associated with an in-creased risk for developing bladder, head, larynx, breast, skin, colon, stomach, lung, and testicle cancer [20] [25] [30] -[32] .
Upon examination of our results for each gene individually, we found a significant difference of frequencies for GSTM1 between the two groups, since the wild type allele had a greater presence in the breast cancer group than in the control group, these results are similar to those obtained by Zheng et al. (2003), in a study of 326 patients with this neoplasm, finding an association between wild GSTM1 and alcohol consumption with susceptibility to breast cancer [33] . However, these results are contrary to expectations, since an individual with a GSTM1-null polymorphism, would be expected to have an increased susceptibility to cancer, as demonstrated by Garcia et al. (1999). Studying a group of 466 women with breast cancer, they conclude that GSTM1-null increases the risk of developing this malignancy [34] .
Regarding the frequencies of the GSTT1 genotypes, no significant difference between the groups was found, these results agree with those obtained in previous studies, one of them by Hashemi et al. (2012), on 134 patients with breast cancer and another by Vogl et al. (2004), on 2048 patients, no association between GSTT1 genotypes and the presence of breast cancer [35] [36] was found in either study. This is in contrast with the findings of Garcia et al. (1999), who in studying this gene concluded that GSTT1null genotype substantially increases the risk of developing this neoplasm [34] .
Sohail et al. (2013) conducted a study on 100 patients diagnosed with breast cancer and Lu et al. (2011) conducted a meta-analysis of 34,658 patients; both studies found an increased susceptibility of heterozygote genotypes for breast cancer [37] [38] . On the other side, studies by Vogl et al. (2004), Hashemi et al. (2012), as well as the present investigation found no relationship between GSTP1b genotypes with breast cancer [35] [36] .
Particularly striking is the absence of information in literature on the involvement of GSTP1 exon 6 polymorphisms with breast cancer. Our results show significant difference for this polymorphism between patients and the control group, from which we infer a possibility that the heterozygote polymorphism (P1a/P1c), may be associated with breast cancer. This seems inviting to continue studies on this gene.
Previous studies have explored susceptibility on a single gene, taken individually; joint participation, the way it occurs in vivo, has not been reported. Far from finding the variants corresponding to deletions (GSTM1-null
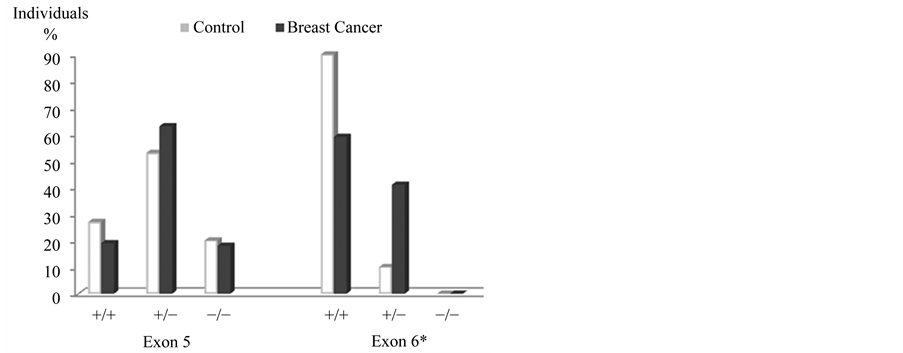
Figure 2. Frequencies of GSTP1 polymorphisms, exon five and six. *Significant differences in GSTP1, exon 6: wild P1a/P1a and heterozygous GSTP1b P1a/P1c, p ≤ 0.05.

Table 2. Frequencies for GSTM1, GSTT1 and GSTP1 genotypes by group.
and GSTT1-null), and mutations in exon 5 (GSTP1b) and exon 6 (GSTP1c), as could be expected in the group with breast cancer, we found that the genotype with GSTM1-wild, GSTT1-wild, exon five GSTP1 homozygote (P1b/P1b) and exon 6 (P1a/P1a) wild was the most frequent in the group with breast cancer, while the most frequent genotype in the control group was that with GSTM1-wild, GSTT1-wild, exon five GSTP1 heterozygote (P1a/P1b) and exon six (P1a/P1a) wild. Given the preferential presence of wild genotypes in both groups, our results do not allow us to infer that these polymorphisms confer an increased risk of developing breast cancer. Similar results were obtained by Vogl et al. (2004) [35] .
5. Conclusions
Studies such as this invite to document triggering or precipitating factors such as habits, habitat and work activities, among others, to have a more comprehensive picture of cancer development.
The different combinations of gene polymorphisms reported here showed no association with the development of breast cancer.
Acknowledgements
Extended to Dr. José Luis Barrera and Dr. Paula Cabrera ISSEMyM State Oncology Center and the Adolfo Lopez Mateos State Medical Center in Toluca, México, for their support in obtaining the samples. To all participants in this study. To Dr. Eduardo Ramirez San Juan for his advice on data analysis. This project was partially funded by the UAEMéx, agreement No. 3452/2013CHT.
NOTES
*Corresponding author.