QF-PCR as a molecular-based method for autosomal aneuploidies detection ()
1. INTRODUCTION
Autosomal aneuploidies that allow survival to fullterm, namely, trisomy 21, 18, and 13 account for 89% of chromosome abnormalities with a severe phenotype [1,2]. Since 1970, the routine using of karyotyping has been regarded as the gold standard for prenatal diagnosis of chromosomal abnormalities. It has the advantage of detecting not only numerical chromosomal abnormalities, but also structural chromosomal rearrangements [3,4]. However, conventional cytogenetic analysis necessitates the culture of prenatal material obtained through amniocentesis or chorionic villous sampling (CVS). The unavoidable delay in reporting with subsequent postponement in decision-making regarding continuation of the pregnancy considerably increases parental anxiety.
Quantitative Fluorescent Polymerase Chain Reaction (QF-PCR) has emerged as a rapid and cost-efficient prenatal diagnostic test for selected chromosome aneuploidies [5]. QF-PCR tests are now performed in several prenatal centres in Europe for the detection of major numerical abnormalities affecting chromosomes 21, 18, 13, X and Y, with results provided within a 24 h period [6-13]. Despite the wide range of microsatellite marker multiplexes used by these laboratories, the assays are reported as both robust and reliable [4].
In this study, we report our experience concerning the performance of QF-PCR as a molecular technique for the detection of chromosome 21, 18 and 13 copy numbers compared to the gold standard conventional cytogenetic analysis.
2. MATERIALS AND METHODS
This study was performed at the Institute of Medical Genetics and Human Genetics, Charité Universitätsmedizin, Berlin, Germany. One hundred and twenty-one archival prenatal samples, in the form of cultured amniocytes, chorionic villous sample (CVS) long-term cultures and DNA samples prepared from native amniotic fluid, CVS and cord blood, were analysed. Forty-two archival postnatal samples in the form of cultured fibroblasts and heparinised whole blood samples were also included. DNA isolation was performed following standard protocols at the Institute of Medical Genetics and Human Genetics. The genomic DNA extracted from whole blood samples was subsequently purified using the automated BioRobot® M48 (Qiagen, Hilden, Germany) following the manufacturer’s instruction.
The enrolled samples were blindly tested for chromosome 21, 18 and 13 copy numbers using QF-PCR. Subsequently, the results were compared with the original karyotype results obtained from the registry of the institute of Medical Genetics and Human Genetics.
Multiplex PCR using 12 fluorescently labelled primer pairs (Applied Biosystems, Carlsbad, Carlifornia) was applied for co-amplification of 12 microsatellites markers; with four on each of chromosomes 21, 18 and 13. The multiplex contained primers for D21S1435, D21S11, D21S1270, D21S1411, D18S391, D18S978, D18S386, D18S535, D13S742, D13S634, D13S628 and D13S305. The primer combination followed that of Mann et al. [6] with one modification, in that a working primer mix containing all primers at equimolar concentrations (2 µM each primer) was used. PCR was set up in a 25 µL reaction volume containing 12.5 μL 2x Qiagen Multiplex PCR Master Mix (Qiagen), 2 μL working primer mix (160 nM each) and 1 µL template DNA (100 - 200 ng). The PCR cycling conditions for all samples were consistent and performed as previously described by Mann and colleagues [6]. The amplification products were then stored at 4˚C until further analysis. Amplification was carried out using a GeneAmp® PCR System 9700 thermocycler (Applied Biosystems). Fragment analysis of the PCR products was carried out using the 3730 DNA Analyzer 48 Capillary Array, 36 cm (Applied Biosystems) with Data Collection v2.0 software (Applied Biosystems) and finally the GeneMapper® Software v3.7 (Applied Biosystems) for fragment sizing and quantification. Each amplified sample (0.5 µL) was added to 9 µL of Ultrapure Hi-Di Formamide (Applied Biosystems) and 0.2 µL of GeneScan-500 Rox size standard (Applied Biosystems) in a MicroAmp® optical 96-well reaction plate (Applied Biosystems). Prior to electrophoresis, the mixture was denatured for 5 min at 95˚C. Finally, samples were loaded into the 3730 DNA Analyzer and subjected to capillary electrophoresis. A normal and trisomic control sample was included in each run.
Peak area measurements were used to calculate allele ratios. Interpretation of the allele ratios was done in accordance to the professional guidelines for clinical cytogenetics and clinical molecular genetics, QF-PCR for the diagnosis of aneuploidy best practice guidelines (2007) v2.01. In brief, the presence of two alleles with an allele ratio between 0.8 - 1.4 at a given locus was considered to represent a normal diallelic pattern. The presence of either three alleles in a 1:1:1 ratio or two alleles with a ratio of < 0.65 or > 1.8 was considered to represent a trisomic pattern. The presence of only one peak was considered to be non-informative. A minimum of two informative markers was required to provide information for each chromosome. Markers demonstrating allele ratios falling in the intermediate ranges (1.4 - 1.8 and 0.65 - 0.8) were referred to as inconclusive results.
Statistical analysis was performed using IBM SPSS v 19.0. The t-test p-value of < 0.05 was considered statistically significant.
3. RESULTS
The study was performed on 163 samples; 115 autosomal aneuploidy cases, five unbalanced rearrangements affecting chromosome 21 and 18, and 43 controls with normal karyotype. The autosomal aneuploidy group included 67 trisomy 21 cases (61 non-mosaic, five mosaic trisomy 21 samples and one trisomy 21 sample with MCC), 36 trisomy 18 cases (35 non-mosaic and one mosaic trisomy 18 samples) and 12 non-mosaic trisomy 13 cases.
The results of the QF-PCR were in agreement with those of cytogenetic analyses in 113 out of 120 total abnormalities tested, whereas in the remaining seven samples, the QF-PCR results were either inconclusive or normal (Table 1). All 43 normal samples were successfully assigned as normal for chromosomes 21, 18 and 13 copy numbers with QF-PCR. Sensitivity was 94.2%, whereas specificity was 100%. Considering only nonmosaic trisomies (n = 108), there was no discrepancy between the QF-PCR and karyotype results in all samples, reflecting a sensitivity of 100%. Multiplex marker assay was informative in 100% of samples with at least two informative markers for each autosome tested. In 64.8% of non-mosaic trisomy samples (70 samples), the diagnosis was based on the results of four abnormal markers. In 31.5% (34 samples), diagnosis was based on three abnormal marker results while the fourth marker was uninformative. In 3.7% (four samples), diagnosis was based on the results of two abnormal markers while the other two markers were uninformative.
Marker heterozygosity was calculated in the study sample based on the frequency of each marker being informative. The calculated heterozygosities compared to those reported by Mann et al. [14,15] are listed in Table 2. The observed allele size ranges, reported in bp, are shown in Table 3 compared to those of Mann et al. [6].
Mosaicism was considered in cases showing consistently minor extra allele peaks or inconclusive allele ratios falling in the intermediate ranges on a chromosomespecific group of markers [16]. One mosaic trisomy 21
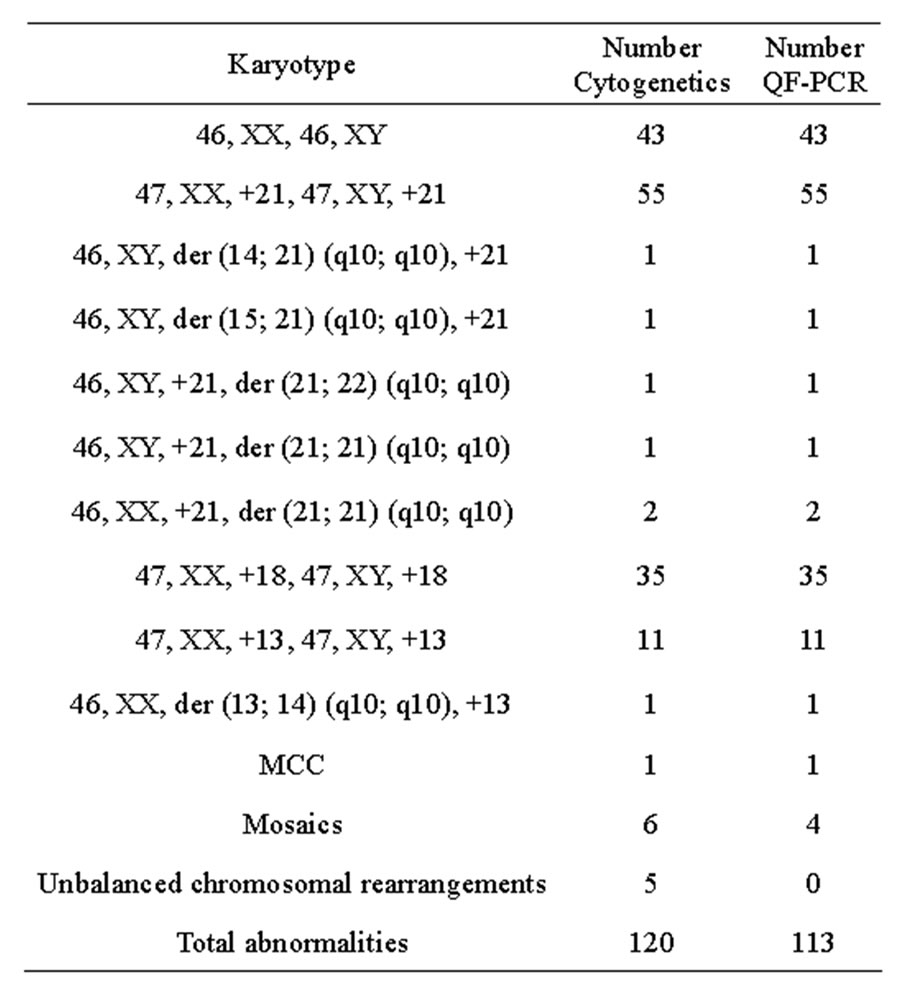
Table 1. Comparison of the results of testing 163 samples for chromosome 21, 18 and 13 copy numbers using QF-PCR with cytogenetic results. Karyotypes are described according to the ISCN 2009 nomenclature.
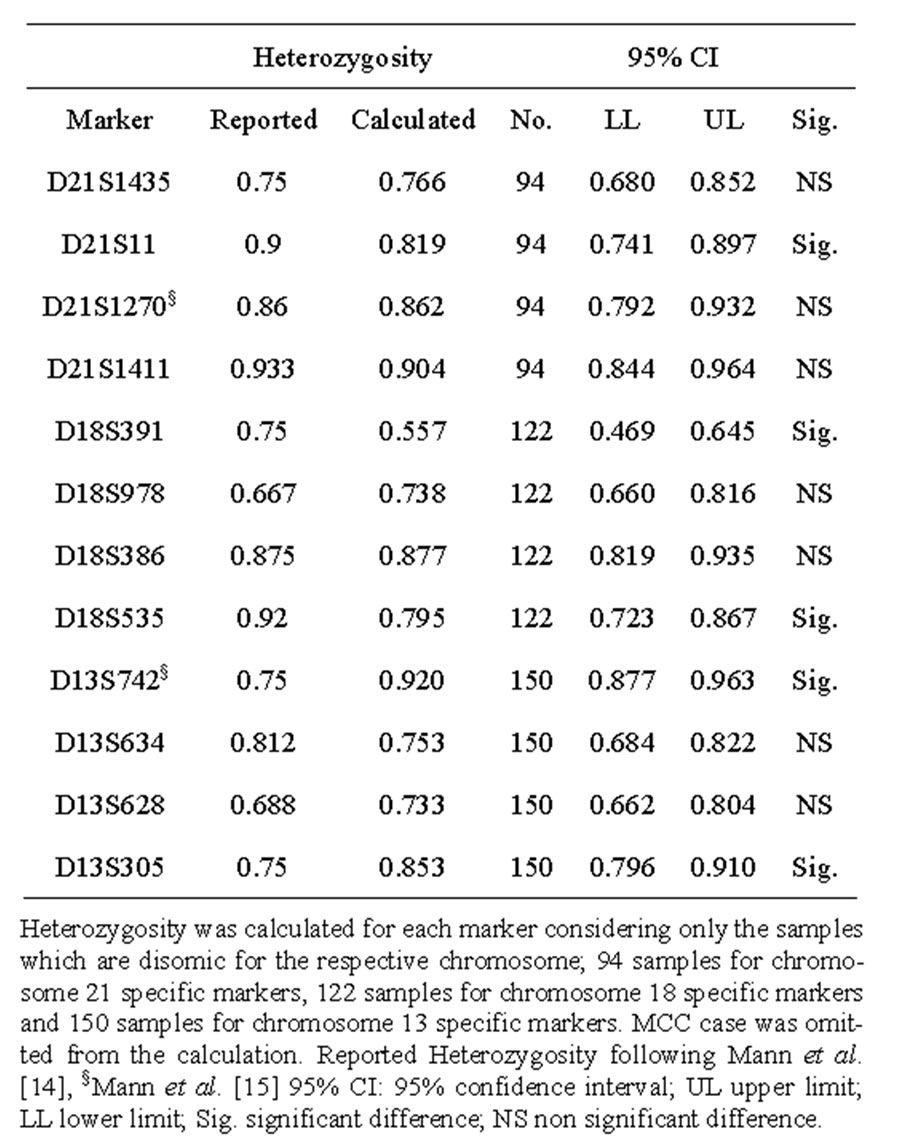
Table 2. Calculated heterozygosity of the Markers included in the autosomal chromosome multiplex compared to those of Mann et al.
case with 92.0% trisomic cells: mos47, XY, +21 [46]/46, XY [4] was successfully assigned by QF-PCR demonstrating abnormal diallelic results for all chromosome 21 markers. Three samples were recognized by QF-PCR as possible mosaics for trisomy 21, and this was confirmed by the karyotype results. Two of them were found to be mosaic trisomy 21 with 23% trisomic cells: mos47, XX, +21 [14]/46, XX [46]. Both samples, which were CVS and amniotic fluid from the same patient, demonstrated inconclusive diallelic ratios for two informative chromosome 21 markers, namely, D21S1435 and D21S- 1270. The third case was recognized to be mosaic trisomy 21 with 6% trisomic cells; however, mosaicism was evident as inconclusive allele ratios for two chromosome 21 informative markers, namely, D21S1270 and D21S1411. QF-PCR was unable to detect two cases of mosaic trisomies; one mosaic trisomy 21 with 6% trisomic cells: mos47, XX, +21 [3]/46, XX [47] and one mosaic trisomy 18 with 10% trisomic cells: mos47, XX +18 [5]/46, XX [45]. All informative markers showed normal diallelic results in both samples.
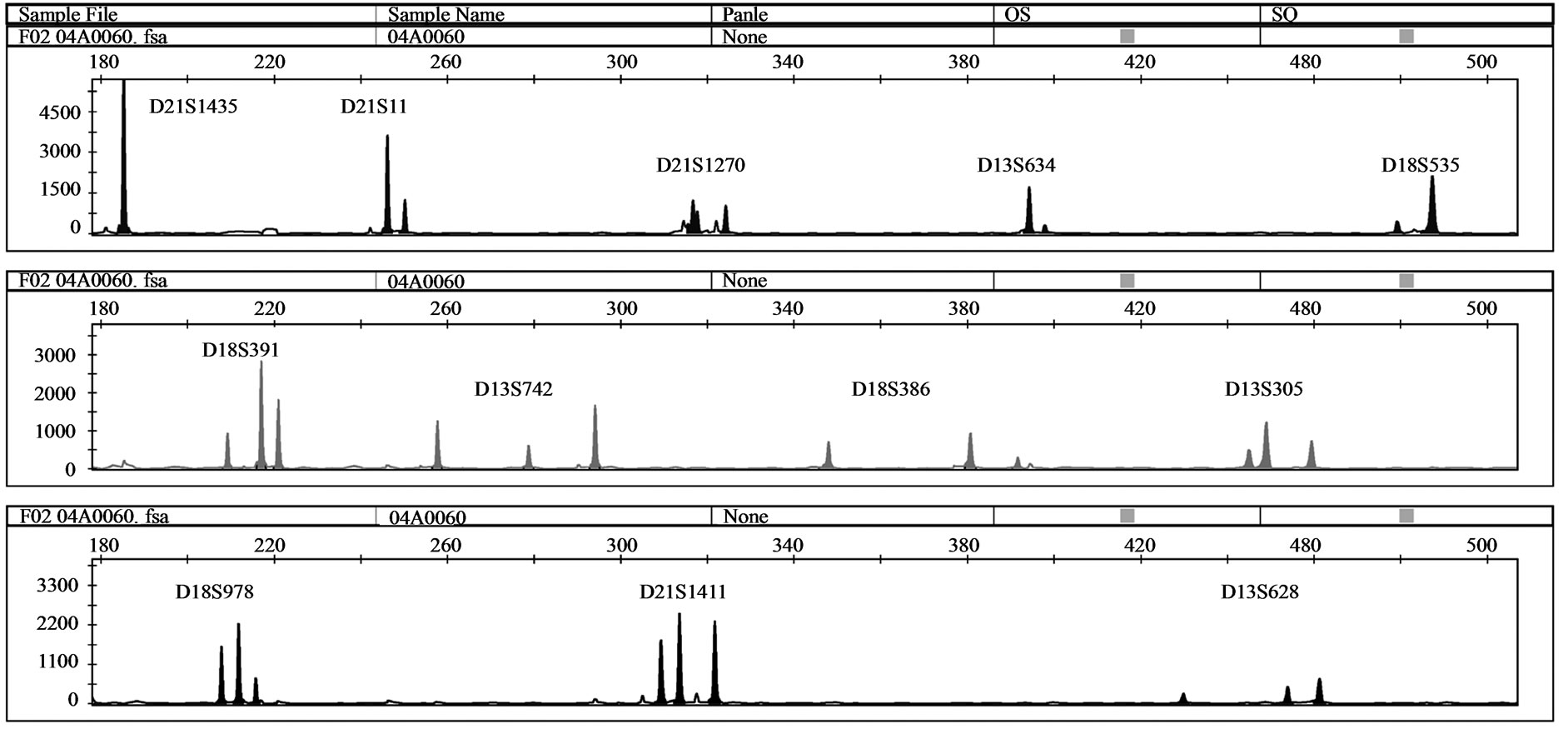
One MCC sample was detected by QF-PCR. The characteristic MCC allele pattern consisted of inconclusive diallelic results, and in case of triallelic results, the peak areas of the maternal-specific and fetal-specific alleles equalled the area of the shared maternal-fetal allele (Figure 1) [16]. The sample was found to be a trisomy 21 case with MCC: 47, XY, + 21 [17]/46, XX [1] (~ 6% MCC) as evidenced by the original karyotype results. In all 39 analysed DNA samples extracted from native amniotic fluid and CVS, no MCC could be detected.
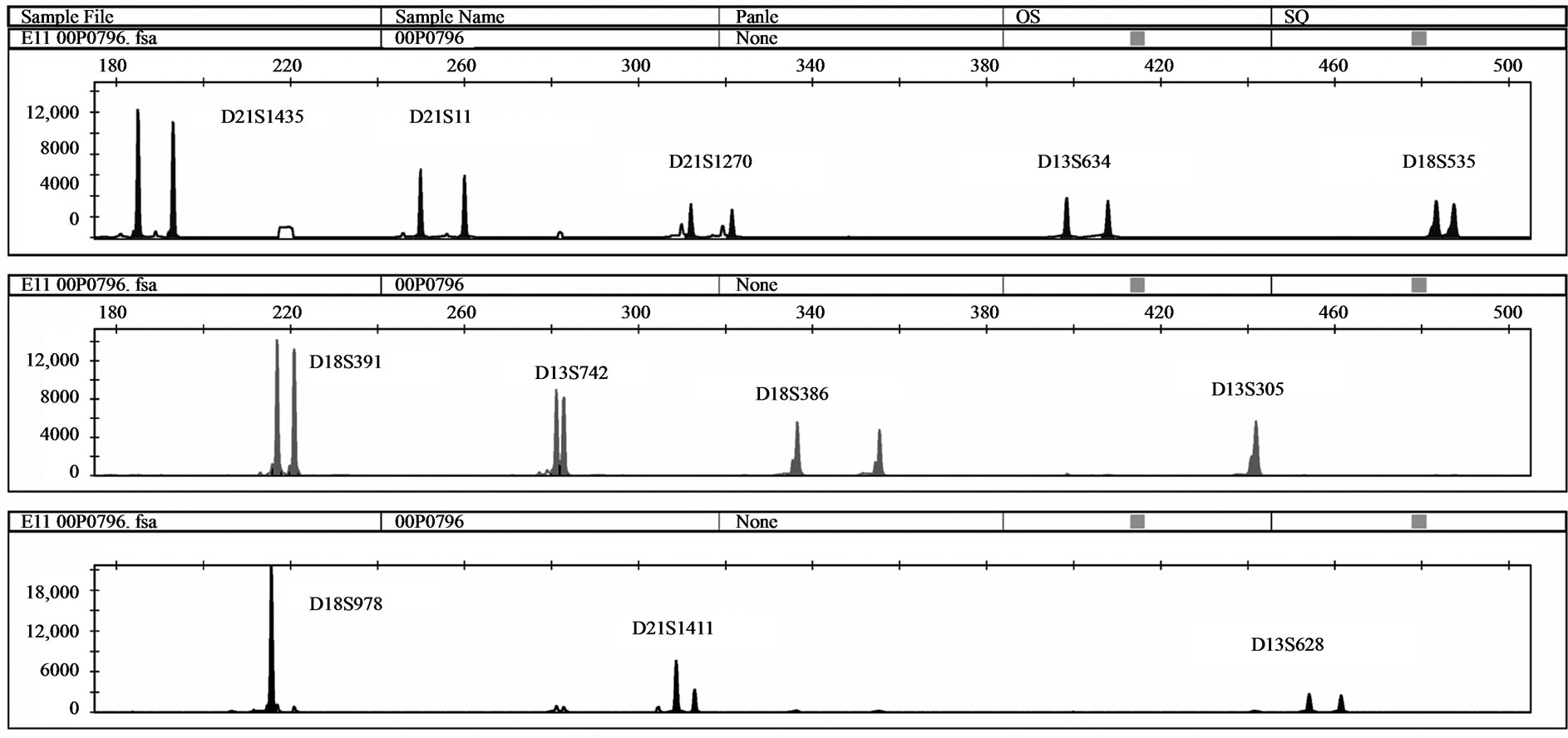
Five samples with unbalanced autosomal rearrangements were tested with QF-PCR. Details for the samples are listed in Table 4. In sample No. 1, QF-PCR demonstrated evidence of partial chromosomal imbalance for chromosome 21 in form of a trisomic diallelic result for D21S1411, while all other informative chromosome 21 markers were normal (Figure 2(a)). The sample was found to be an unbalanced translocation of the long arm of chromosomes 21 onto chromosome 18 leading to partial trisomy 21 and partial monosomy 18. All informative chromosome 18 markers showed normal diallelic pattern. Sample No. 2 demonstrated a QF-PCR profile in which one chromosome 21 marker (D21S1270) showed normal diallelic pattern, whereas the other three markers were uninformative; D21S11, D21S1435 and D21S1411. (Figure 2(b)). The probability of this occurring by chance alone is low based on the previously experienced high heterozygosity of the markers included in the multiplex assay. The sample was found to be partial monosomy 21, partial trisomy 22. Both D21S1435 and D21S11 are located in the monosomic region. QF-PCR Marker assay was normal in two samples with a partial chromosome 18 deletion (samples No. 3 & 4). One partial mosaic tri
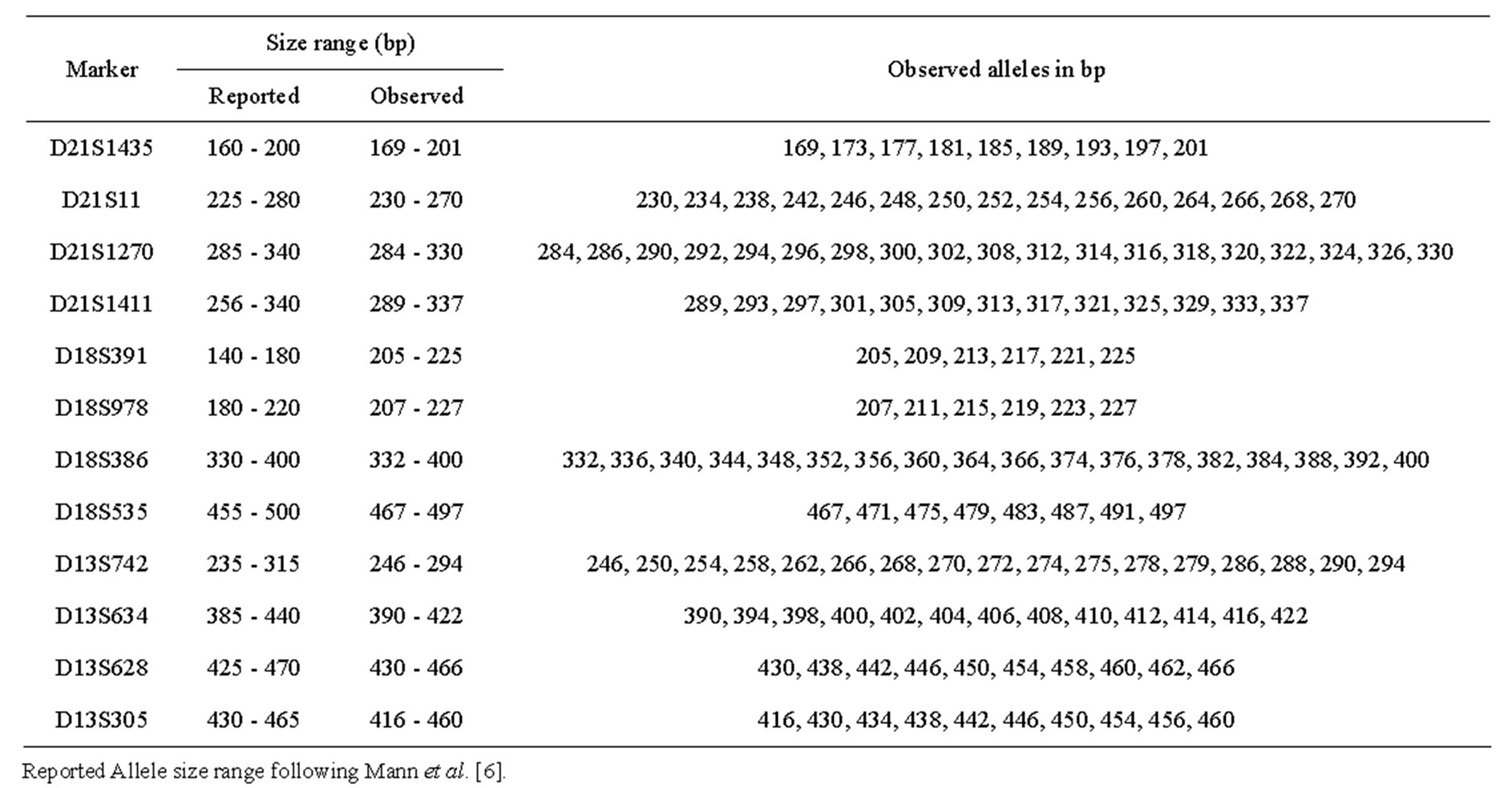
Table 3. Allele size range of the microsatellite markers included in autosomal chromosome multiplex compared to those reported by Mann et al. together with observed alleles in bp.
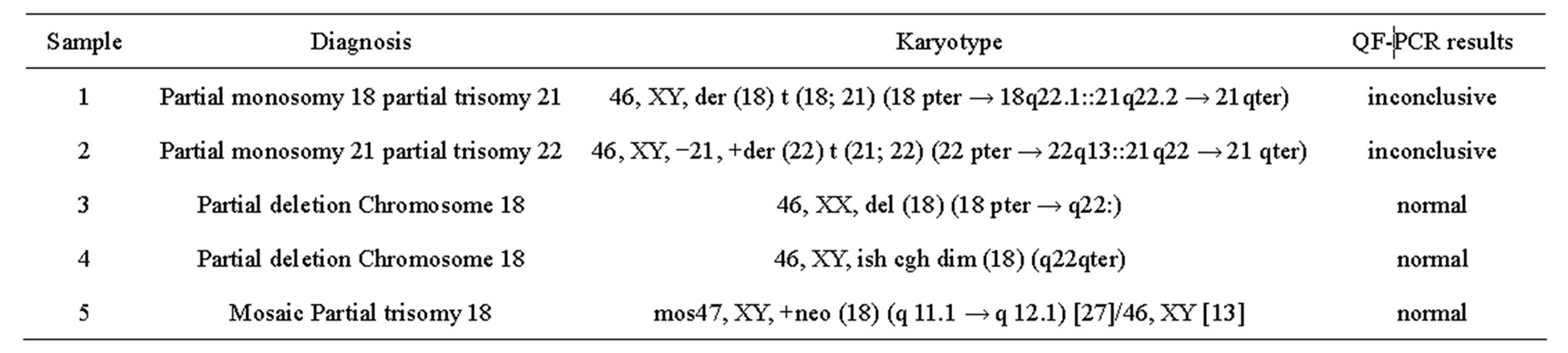
Table 4. QF-PCR results of the unbalanced autosomal rearrangements.
somy 18 sample was tested and demonstrated normal results for all chromosome 18 markers (sample No. 5).
4. DISCUSSION
Using QF-PCR, a sensitivity of 94.2% and specificity of 100% was demonstrated. Our results were comparable to those reported in the literature [9,10]. All seven cases which could not be confirmed by the molecular assay were either mosaic trisomy samples with a low level of the trisomic cell line (two cases) or unbalanced structural chromosomal rearrangements (five cases). Considering only the non-mosaic trisomy cases, the multiplex performance reflected 100% sensitivity and specificity. These results were in agreement with published data [6,10,15, 17,18].
Multiplex marker assay was informative in 100% of samples with at least two informative markers for each autosome tested. No additional markers were required.
This could be attributed to the high heterozygosity ratios observed in our study. With the exception of D21S11, D18S391 and D18S535, whose calculated heterozygosity ratios were significantly lower than those reported by Mann and co-workers, [14] our calculated heterozygosity ratios were either significantly higher or showed nonsignificant difference to those reported. With the exception of D18S391, our observed size ranges of the markers used were more or less close to those reported by Mann and colleagues [6]. The observed size range of D18S391 consistently appeared to be different to the reported one. However, this caused no interpretational problem due to the unique allele size/label colour combination.
Using QF-PCR, four out of the six mosaic trisomy samples could be detected. The lower limit for detection in mosaic samples was 23% trisomic cell line. This was in agreement with Mann et al. [6] and Cirigliano et al. [10] who concluded that cell lines contributing at least 20% of the total cell population can be confidently iden-
 (a)
(a)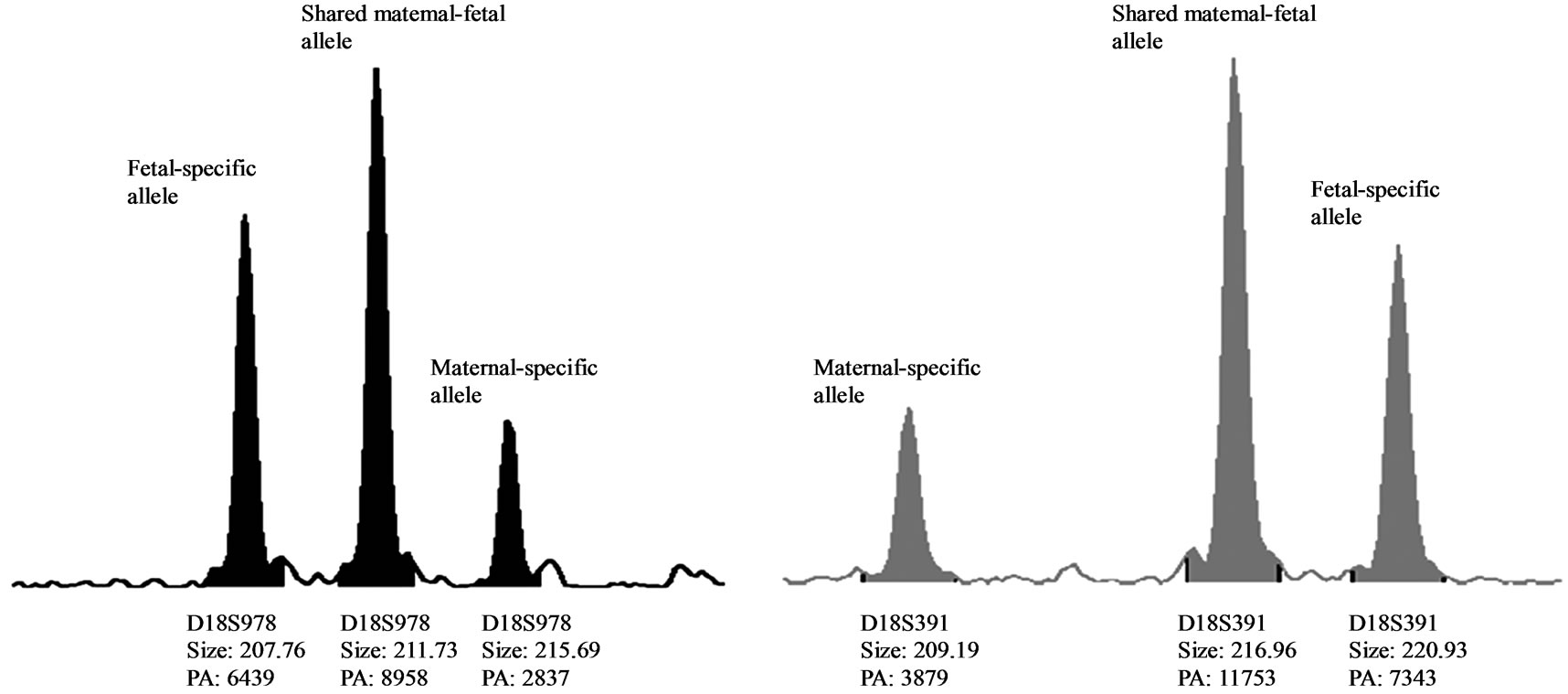 (b)
(b)
Figure 1. (a) QF-PCR electrophoretogram of a trisomy 21 case showing MCC. A third small maternal-specific allele can be observed for D21S1270, D18S391, D13S742, D18S386, D13S305, D18S978, D21S1411 and D13S628. D21S11, D13S634 and D18S535 show inconsistent diallelic results. D21S1435 is uninformative. (b) & (c): A closer view of D18S978 and D18S391 showing evidence of maternal cell contamination. The peak areas of the maternal-specific and fetal-specific alleles almost equalled the area of the shared maternal–fetal allele. The primer labelling is 6-Fam, HEX and NED (from top to buttom). PA: Peak Area.
tified. However, a single mosaic trisomy 21 case with only 6% trisomic metaphases in the conventional cytogenetic analysis was identified by QF-PCR: mos47, XX, +21 [3]/46, XX [47]. The estimated level of trisomic genotype by QF-PCR was approximately 30%. This contradiction could be attributed to the fact that different cell types were used for both tests. While the conventional cytogenetic analysis of peripheral blood is based on PHA stimulated T-lymphocytes, for QF-PCR DNA from whole blood is used which contains all white blood cells with a high percentage of granulocytes. Our results therefore suggest that the percentage of trisomic cells was higher in the cell fractions others than the T-lymphocytes. FISH could not be performed for confirmation of the mosaic percentage due to the lack of native sample material suitable for testing by FISH.
In this study, maternal cell contamination could be detected in a single trisomy 21 sample. The original cytogenetic results reported one out of 17 cells counted as maternal: 47, XY, +21 [17]/46, XX [1] (~ 6% MCC). The estimated level of maternal genotype by QF-PCR was approximately 30%. The level of MCC was too high
 (a)
(a)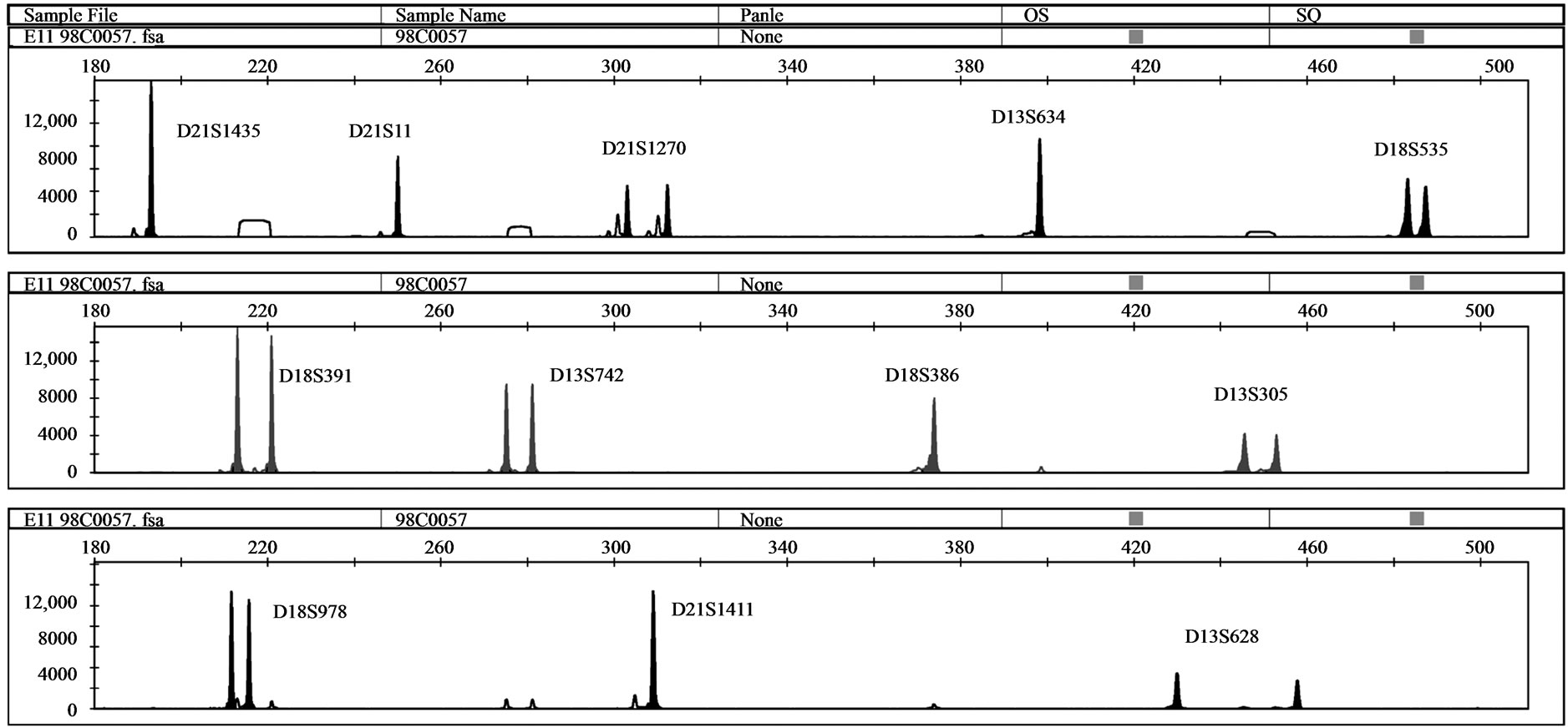 (b)
(b)
Figure 2. (a) QF-PCR electrophoretogram of a partial trisomy 21 partial monosomy 18 case displays a trisomic diallelic pattern for D21S1411 (allele ratio: 2:1), whereas all other informative chromosome 21 and 18 markers are normal (allele ratio: 1:1); (b) QF-PCR electrophoretogram of a partial Monosomy 21 case. D21S1435, D21S11 and D21S1411 are uninformative.
to allow for detection of the fetal trisomy genotype by QF-PCR. The accurate percentage of MCC could not be clarified as the original sample material was not subjected to I-FISH assay.
QF-PCR was able to suggest rather than diagnose two unbalanced autosomal rearrangements affecting chromosomal segments harbouring one or more markers of the multiplex assay. In the three other cases, normal QFPCR assay patterns were obtained due to the location of the tested markers outside of the rearranged region. As the molecular assay is not designed to detect all chromosomal abnormalities, missing of such unbalanced chromosomal rearrangements was expected. One case with partial monosomy 18 partial trisomy 21 was of special note; the QF-PCR pattern was consistent with partial chromosome 21 imbalance evidenced by trisomic results for one chromosome 21 marker while all other markers were normal. As the same pattern could also be due to rare submicroscopic polymorphic duplications of the microsatellite, analysis of samples from both parents with the same microsatellite marker is mandatory in such cases in order to distinguish both possibilities. Submicroscopic polymorphic duplications of microsatellites are primarily inherited and have no clinical significance [6,10]. This would also provide a warning about the risk of reporting an autosomal trisomy detected with a single abnormal marker [10].
Our amplification failure rate was zero percent in QFPCR on DNA extracted from cell cultures. The multiplex PCR showed consistent results with the initial DNA amount of 100 - 200 ng/reaction. Our DNA extraction method was able to produce an adequate amount of genomic DNA even with low cellular concentration. Apart from the postnatal heparinised whole blood samples, the quality of DNA was suitable for an efficient multiplex PCR. Postnatal heparinised whole blood samples failed to amplify upon initial experimentation. This could be explained by the inhibitory effect of residual heparin on multiplex PCR. Following re-extraction using an automated method, all samples demonstrated successful multiplex PCR results. Lower concentrations of DNA could also be tested, provided that the amount of sample loaded for sequencing was modified accordingly. QF-PCR is reported to generally have a low amplification failure rate [6, 10]. The QF-PCR method showed excellent reproducibility. A normal control sample was consistently analysed with each run, with no remarkable inter-run variation in peak area, size or allele ratios. Testing different sample types for the same case (fetal blood vs. amniotic fluid; CVS vs. amniotic fluid) demonstrated consistent results which were almost identical in all parameters; peak size, peak area and allele ratios.
QF-PCR demonstrated great efficiency. QF-PCR correctly identified all the non-mosaic numerical abnormalities related to chromosomes 13, 18 and 21, i.e. 100% sensitivity. QF-PCR showed 100% specificity. QF-PCR was able to identify mosaics with 23% trisomic cells or more. QF-PCR was able to identify MCC. As QF-PCR is primarily designed to detect selected chromosomal disorders, it is at risk of missing unbalanced chromosomal rearrangements for which they are not designed.
5. ACKNOWLEDGEMENTS
This work was done in the context of a DAAD-German Egyptian Research Long-Term Schoolarship (GERLS).