1. Introduction
Marine bacteriophages, in spite of their ubiquitous occurrence in both salt and fresh water environments have only been extensively studied for about thirty years. As a consequence our understanding of their architectural features is substantially less than their terrestrial counterparts. The ecology of marine bacteria, however, is different, and we might anticipate that the corresponding phages would exhibit novel structural features to fulfill functions not commonly required of terrestrial phages. To this point, however, no substantial structural components have been reported for marine bacteriophages that set them apart from the phages with which we are familiar. We describe here the observation of perhaps the first significant new structural element displayed by some marine bacteriophages that have not, otherwise, been seen, and we suggest a possible function.
Most tailed bacteriophages engage their hosts through interaction of the termini of segmented tail fibers with the outer membrane of susceptible cells. The termini possess recognition properties and the lengths of the tail fibers more or less determine the search space of the virus. For terrestrial phages the search space need not be great because host cells grow in colonies or at high densities, the distance between cells is relatively small, and infection spreads easily. This is frequently not the case in marine environments. Many bacteriophages also have shorter fibers, referred to whiskers that originate at the phage collars, the junction between the icosahedral head and the tail assembly. The function of these is more obscure, but they presumably supplement the recognition functions of the long tail fibers. The novel feature that we have observed is very likely homologous to one or both of those fiber components.
2. Materials and Methods
Seawater samples yielding phage characterized by the 3 + 1 fiber tassel pattern were drawn in July 2008 (SCAM-4) from the Pacific Ocean coastal waters surrounding Balboa Island (Newport Beach, CA) and from Seal Beach, CA (S-CAM-3) in August 2008. The isolate containing the phage exhibiting the 5 + 0 tassel pattern was obtained in February 2008 from the waters off Seal Beach, CA (S-CAM-1). The viruses were isolated on Synechococcus sp. WH7803 by dilution-to-extinction in 48-well microtiter plates. A lysate from one well was further purified by two rounds of plaque selection on soft agar plates. In coastal waters of southern California, Synechococcus spp. commonly reach concentrations above 105 cells/ml. Cyanophage titers infecting WH7803 reach over 103 ml−1 of seawater during the summer and often drop below 101 ml−1 in the winter and spring.
AFM analyses were generally carried out in air, though in some cases scanning was performed in media. The substrate was freshly cleaved mica or mica coated with poly-l-lysine. Most details of the AFM methods have been presented in earlier papers [1-4]. Media samples of 1.5 µl to 5 µl containing virus were applied to the substrate and the virus allowed to sediment for 15 min to 30 min. The sample, on the substrate, was then exposed to 5% glutaraldehyde in buffer for 2 minutes and excess glutaraldehyde solution shaken off. The substrate was then washed two times with distilled water and dried in a stream of dry nitrogen gas. Alternatively, the sample, after sedimentation, was simply allowed to dry in air onto the substrate after excess liquid was removed on filter paper, and then fixed with glutaraldehyde. The glutaraldehyde is necessary in most cases to rigidify biological samples sufficiently that they permit scanning at high magnification.
AFM imaging was carried out using a Nanoscope III multimode instrument (Veeco Instruments, Santa Barbara, CA). When scanned in liquids, virus and associated macromolecules were scanned at 26˚C using oxidesharpened silicon nitride tips in a 75 ml fluid cell containing buffer. For scanning in air, silicon tips were employed. The images were collected in tapping mode [5,6] with an oscillation frequency of 9.2 kHz in fluid and 300 kHz in air, with a scan frequency of 1 Hz. Vertical and lateral distances were calibrated using standards from Digital Instruments (Santa Barbara, CA) of 10 nm height steps of pitch 200 nm, and standards from MCNC Analytical Labs with step height 10 nm and pitch of 1 um. Cantilevers for scanning in air were from Veeco (Santa Barbara, CA) and were TAP150 with lengths of 115 - 135 um and spring constants of 5 Nn. For solution scanning, they were 0TR4 with lengths of 100 um and spring constants of 0.08 Nn.
In the AFM images presented here, height above substrate is indicated by increasingly lighter color. Thus points very close to the substrate are dark and those well above the substrate white. Because lateral distances are distorted due to an AFM image being the convolution of the cantilever tip shape with the surface features scanned, quantitative measures of size were based either on heights above the substrate, or on center to center distances on surfaces.
3. Results
In the course of our atomic force microscopy (AFM) analyses of bacteriophages infecting cyanobacteria, in particular synechococcus, we encountered an otherwise unexceptional virus that exhibited a novel, and so far as we know, unreported structural feature. Some examples of the bacteriophage are shown in Figure 1. The phage, not unlike others we have observed, has a T = 7 icosahedral head of diameter about 80 nm, as determined by direct visualization with AFM, and a tail assembly about 175 nm in length. Overall the structure is similar to most myoviruses [7], though its base plate is small relative to bacteriophage T4 and its tail assembly longer [8]. Both short spikes and conventional, long tail fibers adorn the base plate, features also common to terrestrial bacteriophages such as T4 [9] and some marine phages [3].
What are unique about this cyanophage, and which is illustrated by the AFM images in Figure 2, are long flexible fibers that originate at either the base plate, or from the collar of the phage. There are never more than two such fibers emanating from either of the two sites, but on some phages, fibers emerge from both origins. These are the two origin points of tail fibers and whiskers [8,10] on those terrestrial and marine phages that have been observed to exhibit such appendages. The lengths of the two fibers are always the same at about 450 nm. As is evident in the AFM images of Figure 1, and even more so in the higher magnification images of Figure 2, the fibers are not simply naked threads, but are decorated in a regular way near their distal ends by tassels composed of globular masses, or bulbs. The number and distribution of these masses are not arbitrary, but are strictly according to one of two patterns.
In several plaque purified cyanophage samples (SCAM-3 and S-CAM-4) there is a distinctive 3 + 1 pattern of four bulbs that terminate the threads. The terminal

Figure 1. In (a) the AFM substrate is cluttered with numerous cyanophages exhibiting extended tail fibers terminating in a characteristic tassel of 3 + 1 bulb shaped particles (referred to here as 3 + 1 tassels, or 3 + 1 fibers). In (b) is a phage about to engage with a cell fragment. The phage has two 3 + 1 fibers emanating from its collar where the tail assembly sheath couples to the icosahedral head, and one 3 + 1 fiber originating from its base plate. In (c) is a less common (in our samples) variant where the tassel is composed of 5 consecutive bulbs (5 + 0 tassels or fibers). Plaque purified samples of phage were either all 3 + 1 or 5 + 0 tasselated, suggesting them to represent two distinct strains of the phage. Note that in (c) the phage also displays the tail fibers usually seen on cyanophages as well as many terrestrial phages. Scan areas are (a) 2 um × 3 um, (b) 1.2 um × 1.5 um and (c) 0.5 um × 1 um.

Figure 2. In (a) through (e) are AFM images of individual phage particles that exhibit the 3 + 1 tail fibers, most originating from base plates. In (d) the phage head has burst under osmotic shock to release a bolus of DNA, but the 3 + 1 fiber remains intact. In (f) is a higher magnification AFM image of a 5 + 0 tassel that is just beginning to disintegrate and display some degree of substructure. Scan areas are (a) 1 um × 1 um, (b) 1 um × 1 um, (c) 1 um × 1 um, (d) 0.5 × 0.5 um, (e) 1 um × 1um and (f) 0.5 um × 0.5 um.
three bulbs are immediately adjacent to one another, and possibly in contact, while the forth bulb is always found about 135 nm further upstream toward the phage base plate or collar. The fiber does not pass through the center, or even the interior of the globular units, but is attached at the periphery of the bulbs.
The bulbs, based on AFM height measurements, have a diameter when dry of about 25 nm, but drying produces at least a 25% shrinkage [1,2,4], so the bulbs are probably closer to 30 - 35 nm in diameter. Assuming the globular masses to be composed of protein, this would suggest a total molecular weight of about 5 × 106 D, or about the same as a T = 3 icosahedral plant virus.
As shown by Figure 3, the globular masses are not monolithic, but are themselves composed of many smaller, presumably protein subunits of more or less similar, if not the same sizes. The similarity in subunit size, probably about 25 to 30 kD in molecular weight, based on their measured diameters of around 4 to 5 nm, suggest some sort of defined structure, though we cannot detect any geometric order or pattern at this resolution. As seen in Figure 4, the globular masses sometimes break apart and become unattached to the long fibers, dissociating into constituent subunits. We do not observe there to be any central core or internal material enclosed in the interior of the globular units.
An alternate form of the tassel was observed in another plaque purified phage sample (S-CAM-1) that does not terminate in the characteristic 3 + 1 pattern, but in five consecutive bulbs. Examples appear in Figures 1(c) and 2(f). In this form the bulbs appear otherwise identical to those seen in the 3 + 1 fibers, in both size and composi-
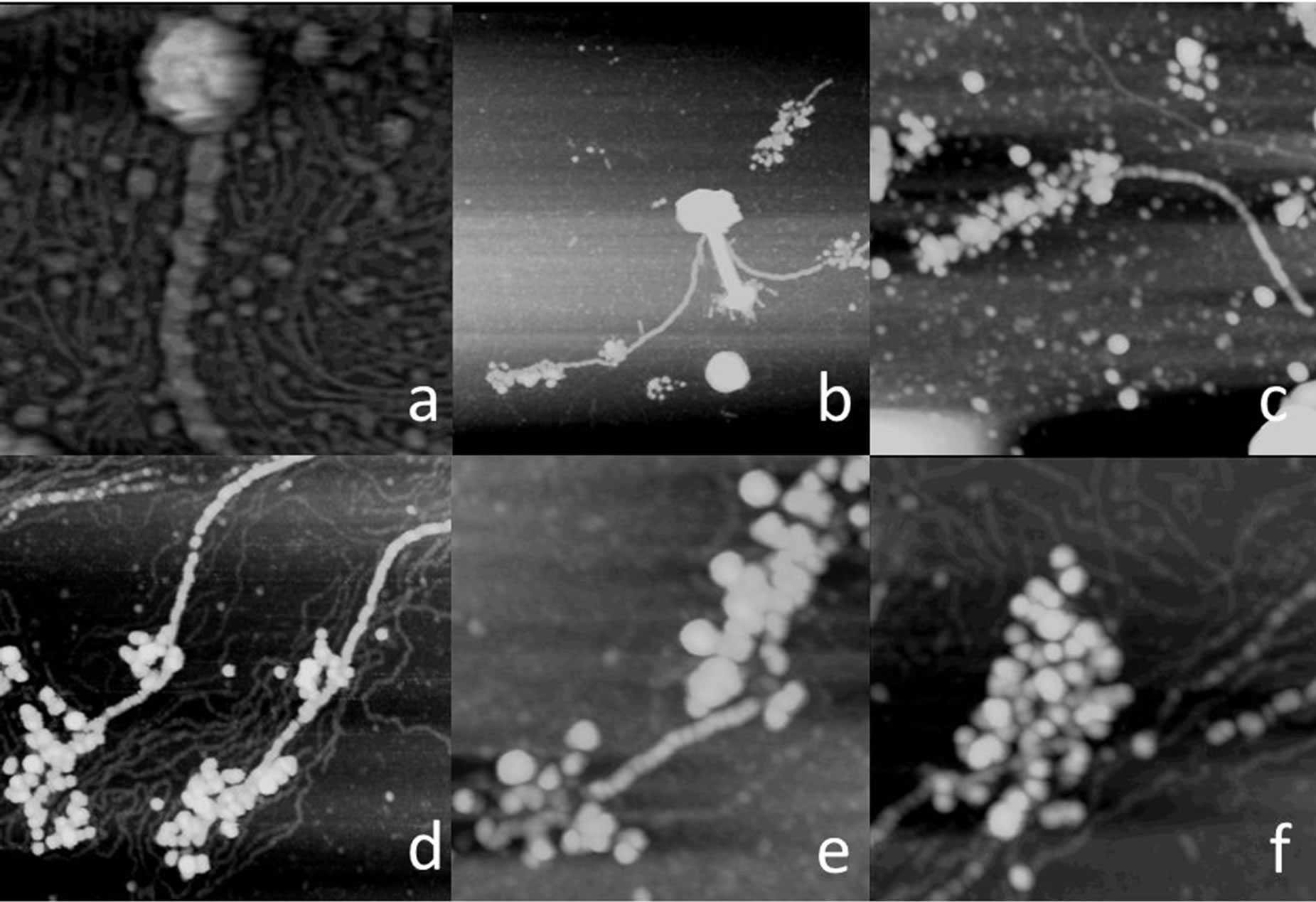
Figure 3. In (a) is the +1 bulb of a 3 + 1 tassel. It is virtually spherical but otherwise almost featureless with no indications of substructure. In (b) through (f) the bulbs are spontaneously disintegrating to reveal themselves composed of subunits of more or less uniform diameter that we presume to be globular proteins. The flexible, much smaller fibers that crowd the background of (a), (d) and (f) are DNA from degraded phage heads. Scan areas are (a) 250 nm × 250 nm, (b) 1 um × 1um, (c) 0.5 um × 0.5 um, (d) 0.5 um × 0.5 um, (e) 250 nm × 250 nm and (f) 250 nm × 250 nm.

Figure 4. The fiber shafts of both the 3 + 1 and 5 + 0 tassels exhibit a distinctive helix that winds about them with a pitch of about 125 nm. The core of the shaft is also helical but made up of multiple strands, very likely three in number, with a helical pitch of about 25 nm. Thus the fiber appears to be a three stranded thread wrapped about its exterior by a forth, unique thread. A proposed model for the entire helical shaft and tassel of bulbs for a 3 + 1 fiber is shown in (d). The scan areas are (a) 0.5 um × 0.5 um, (b) 0.5 um × 0.5 um and (c) 150 nm × 250 nm.
tion. We refer to those as 5 + 0 patterns. For both the 3 + 1 and 5 + 0 tasseled fibers we frequently see them broken off from phage tails or collars and free in our preparations.
A fiber to which the globular masses are attached is also highly structured and complex in its construction. The fiber, which presents a challenge to the resolution of our AFM, appears to be composed of two kinds of filaments, or possible only one kind of filament that performs two structural roles. Hence it is more a thread than a single fiber. Overall, the thread, as illustrated by Figure 4, appears in our AFM images to be composed of three internal filaments that twist around one another to form a triple helical core having a linear periodicity of about 25 nm. A forth fiber then winds around the internal core triplex, but with a much longer helical period of 100 nm. The average diameter of the dried fiber is 7 - 8 nm, or probably about 10 nm when fully hydrated.
The composition of the fibrils making up the threads is not evident. Most fibrils of this kind, particularly those having the observed flexibility and resilience, are frequently comprised of successive coiled-coil proteins [11, 12] but the greater diameter of these fibrils would also be consistent with other motifs, such as actin or other fibers as are seen composing cellular cytoskeletons. They are unlikely to be formed by a single polypeptide, as that would seem incompatible with a length of 450 nm, nor are they segmented as is seen for the tail fibers in other bacteriophage such as T4 [10].
There is another curious observation that we have made on numerous occasions and with multiple samples that deserves description, though we do not fully understand its implications. The spherical globules of the 3 + 1 and 5 + 0 threads appear to have some affinity for the tail assembly of the phage from which they originate, and sometimes for the tail sheaths of other phage in their local environment.
This affinity is evidenced by the AFM images in Figure 5. The globular units are often seen associated (how firmly we cannot judge) with tail sheathes and always in the same manner. They line up in linear register along one side of the tail assembly. A single tail assembly may accommodate either one or multiple sets of globular masses. In Figure 5(f) for example is a tail assembly with three consecutive globular units from one thread aligned along its length. In Figures 5(a)-(c) are tail assemblies with six globular units bound from two threads, 3 in a line along one side, and 3 in a line along the other.
One explanation for the association of the tassels for the phage tails is simply that the fibers and their tassels have thrashed randomly about the phage particles and ultimately come to rest along the tail sheaths due to weak but nonetheless favorable interactions. That would explain why the tassel of one phage is sometimes seen associated with the tail assemblies of other particles. An alternative possibility is that when new phage are produced in host cells, for efficient packaging purposes, the bulbs associate with their own phage tails, as is observed more commonly, thereby reigning in the fibers, avoiding tangles, and allowing more ordered dispersion from lysed cells.
4. Discussion
We can only speculate what the function of these 3 + 1

Figure 5. In numerous cases, as illustrated by these images, phage are observed on which the 3 + 1 or 5 + 0 tassels interact in a unique way with tail assembly sheaths. The bulbs of a tassel line up in a straight row along one side of a sheath. Often, two series of bulbs from two tassels are observed associated with a single phage tail. Generally the tassels that associate with the tail of a phage originated from that same phage, but that is not always the case. Scan areas are (a) 0.5 um × 0.5 um, (b) 1 um × 1 um, (c) 0.5 um × 0.5 um, (d) 1 um × 1 um, (e) 1 um × 1 um and (f) 400 nm × 500 nm.
and 5 + 0 threads might be, but the fact that they arise from points on the phage, the base plate and the collar, where common tail fibers and whiskers originate on other phage, suggest that they may have a homologous function. If that is the case, then the 3 + 1 (and 5 + 0) threads might have the responsibility of searching the environment and securing attachment to susceptible host bacteria. Presumably then, the bulbs that terminate the threads would have molecular, or structural recognition properties. We might speculate further on these extended appendages, which we see here on marine bacteriophages but which have not been observed, in spite of countless EM studies, on terrestrial phage. In the marine environment cyanobacteria do not generally exist in colonies or aggregates where infection progresses among bacteria within contact, or virtual contact distance of one another. In the marine environment the host cells are free floating, not aggregated, and distance-wise are much further apart. In addition, they are subject to the turbulence of the ocean environment. As a consequence, a phage would likely have to search a much larger volume of space to find and secure an acceptable host. The 450 nm length of these tails fibers, and their thrashing aimlessly about, would provide a spherical search volume of nearly 1 micron diameter, a distinct advantage over the diameters of the search volume provided by the fibers of terrestrial phage.
It is, we believe, significant that these tail appendages were seen on marine bacteriophages and never before on terrestrial viruses. It seems entirely likely that the unique microbial ecology of the sea and the distinctive nature of potential hosts and environmental forces may have promoted the evolution of numerous new structural features not previously seen. Thus we might expect future studies to provide additional new, and perhaps surprising architectural elements having novel functions.