Confirmation of Pearl Millet-Napiergrass Hybrids Using EST-Derived Simple Sequence Repeat (SSR) Markers ()
1. Introduction
Sustainable strategies are needed to develop biofuel systems that have high-energy efficiencies, low food security trade-off risks, and significant environmental conservation components. Because of their ability to efficiently utilize water and nutrients, perennial grasses such as Pearl Millet-Napiergrass hybrids (“PMN”; Pennisetum glaucum [L.] R. Br. × P. purpureum Schumach.) are promising sources of germplasm that can be grown for biomass production on more than 445 million hectares of marginal and abandoned agricultural lands worldwide [1-3]. This can be done without affecting native prairie conservation programs because a vast majority of these marginal grasslands no longer contain native species and can provide a resource towards improved agricultural productivity via low-input, perennial grass cropping systems. The utilization of these marginal and abandoned lands would not impact crop land used to produce corn (Zea mays L.), wheat (Triticum aestivum L.), soybean (Glycine max [L.] Merr.), and other major food crops, but it would enhance: 1) erosion control, soil restoration, and carbon sequestration, 2) wildlife diversity refuges, and 3) low-input conservation agricultural practices [4-6].
Pearl millet and napiergrass hybridize to produce vigorous, robust plants [7]. These hybrids essentially have been overlooked as a perennial biomass crop. They have the potential to combine high yields of tropical perennial grasses such as energycane (Saccharum spp.) and integrated agronomics of large-seeded annual grasses such as sorghum (Sorghum bicolor [L.] Moench). PMN hybrid seed production can exceed 1000 kg ha−1 and approach that of commercial forage sorghum [8]. Because of the large quantity and size of PMN seed produced, it can be directly sown using existing planters and reduce establishment costs compared to vegetatively propagated perennial biomass crops such as energycane or Miscanthus Andersson species. In contrast, most perennial forage grasses that have potential as biomass crops, such as switchgrass (Panicum virgatum L.), are typically planted by broadcasting the seed, and these grasses have establishment problems because of small seed size, seed dormancy issues, slow seedling growth rates, and negative responses to high planting densities [9]. Switchgrass, energycane, and Miscanthus are presently considered the leading candidates as grass bioenergy crops, but none of these species is capable of both direct seeding and high biomass production during the establishment year [10]. PMN, however, is easily established from seed and is capable of producing up to 34 metric tons biomass hectare−1 during the establishment year [11,12]. Intercropping napiergrass with climbing legumes has been reported to increase soil nitrogen, soil carbon, and overall biomass yields [13,14], demonstrating the potential of legumes in providing renewable nutrient sources for PMN hybrids in reduced-input management systems.
Because napiergrass and pearl millet share a common genome, this results in the expression of heterosis in their F1 hybrids [15]. The induced homogeneity and natural heterogeneity of pearl millet and napiergrass, respectively, results in PMN hybrid polycultures and reduces the risk of crop failure and environmental impact inherent in monocultures.
PMN hybrids possess additional attributes beyond that of primary bioenergy or forage production. The natural range of carbohydrate resources in PMN hybrids provide added potential to develop feedstocks optimized for diverse biofuel systems, and biomass fractions not utilized by primary conversion platforms offer numerous opportunities for biorefining high-value co-products [16, 17]. Napiergrass has the highest reported leaf protein content among perennial grasses, and methods optimizing its isolation offer platforms for large-scale production of plant protein-derived bioplastic polymers [18,19]. Incomplete hydrolysis derivatives of lignocellulosic material are common and provide another process-based platform for the production of xylo-(XOS) and cellooligisaccharides (COS) with utility as functional foods, feed additives, and specialty chemical platforms [20,21].
PMN hybrids are unique among energy grasses because they can be used as a “seeded-yet-sterile” feedstock. In this system a large quantity of viable F1 seed are produced from PMN crosses, and these seed produce sterile F1 hybrids [7,22]. These sterile F1 hybrids have sufficient cold tolerance to overwinter and perenniate in regions with temperate climates. Because of this hybrid sterility, seed companies are able to capture the value of PMN varieties due to the elimination of farmer saved seed. This sterility also eliminates the concern of these hybrids becoming invasive weeds.
Methods for confirming PMN hybrids are lacking because the hybrids are difficult to distinguish morphologically from napiergrass. PMN hybrids can be separated from their parents by counting the number of chromosomes in the hybrids and the parents; however, this is a time consuming and laborious process [23]. Because pearl millet is a diploid with 14 chromosomes and napiergrass is a tetraploid with 28 chromosomes, the hybrids should be triploids with 21 chromosomes [23]. Therefore, flow cytometry should be a possible means of identifying the hybrids. By determining the DNA content of the parents and their hybrids, their ploidy levels can be estimated. The genetic diversity of napiergrass has been characterized using RAPDs, AFLPs, and isozymes; a genetic map has not been constructed and molecular tools have not been deployed in breeding programs [24- 26]. Azevedo, et al. [27] confirmed the cross species amplification of microsatellite markers in pearl millet and napiergrass to be approximately 50%, indicating the suitability of using microsatellites as a tool for molecular characterization, parental species identification, and hybrid verification in PMN hybrids. In order to address these issues, the objectives of this research were to: 1) produce a population of pearl millet × napiergrass hybrids; 2) identify phenotypic traits that can be used to verify hybrids; 3) identify PMN hybrids using flow cytometry; and 4) develop simple sequence repeats from expressed sequence tags (EST-SSRs) specific to the parental napiergrass accessions and determine if they are suitable for marker-assisted verification of PMN hybrids.
2. Materials and Methods
2.1. Plant Materials and Hybridization
Two pearl millet cytoplasmic male-sterile A-lines (PI 508273 and PI 599192) and one fertile pearl millet line (PI 564585) from the USDA-National Plant Germplasm System (NPGS) along with one fertile pearl millet accession from the Perennial Grass Breeding Program at Texas A&M University were used as the maternal parents (Table 1). One napiergrass genotype (PEPU09FL01) from the Perennial Grass Breeding Program and the cultivar Merkeron [28] were used as paternal parents (Table 1). Individual pearl millet plants were propagated in 25 mm × 25 mm pots, and individual napiergrass plants were propagated in 3.8 L pots.
To make the controlled hybridizations, pearl millet inflorescences were enclosed in glassine pollination bags
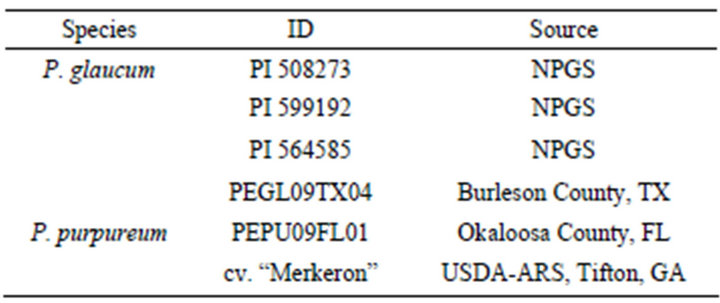
Table 1. Parental species and genotypes used for pollinations and bulked segregant analysis.
prior to stigma exertion to prevent cross-pollination. Stigma exertion from the pearl millet florets was visually confirmed prior to pollination. Napiergrass pollen was collected by gently tapping inflorescences with recently exerted anthers just above an opened 150 mm diameter glass petri dish. Once the pollen had fallen into the bottom half of the dish, it was covered with its top and immediately transported to a flowering pearl millet plant. The glassine bag was removed from the inflorescence, the petri dish cover was removed, and the pearl millet inflorescence, with essentially all of the stigmas exerted from the florets, was gently rolled in the pollen in the bottom of the petri dish. Following pollination, the florets without exerted stigmas were removed using forceps. The inflorescences were then enclosed in glassine bags and remained in the bags until the seed matured. All crosses were made in a greenhouse during the fall of 2010 and winter of 2011 (Table 2).
After the pollinated inflorescences had matured, they were removed from the bags and the seed were threshed, cleaned, and bulked by cross. The two crosses, PI 508273 × Merkeron and PI 599192 × Merkeron, that produced the largest quantity of seed because of the number of pollinations made were separated into four distinct size classes (1.95 mm, 1.81 mm, 1.69 mm, 1.49 mm) using sieves (Seedburo Equipment Co.; Des Plaines, IL). Thirty seed of each size class from these two crosses were used for a germination study. Germination tests were performed across all four seed sizes with three replications. Individual seed from each size class were planted into a commercial potting soil in single compartments (5 cm × 5 cm × 6 cm) in a 30 cm × 30 cm plastic potting flat. Each flat consisted of 36 of the 5 cm × 5 cm × 6 cm cells. The seed were planted at a depth of approximately 1 cm and the flats were placed in a warm greenhouse and regularly watered. The flats were monitored daily for germination for 4 weeks and the percent germination was calculated from these numbers. The seedlings were permitted to grow and were maintained in the greenhouse.
Seed from the other four PMN crosses (PI 599192 × PEPU09FL01, PI 564585 × Merkeron, PEGL09TX04 × Merkeron, and PEGL09TX04 × PEPU09FL01) were germinated as outlined above and grown in the greenhouse. Seedlings from all six PMN crosses were transplanted into a space-planted nursery at College Station, TX on May 10, 2011.
2.2. Phenotypic Traits and Seed Set
After the seedlings were transplanted into a space planted field nursery, each plant was closely examined on a regular basis to determine if it possessed morphological traits of both parents or was similar to the maternal parent. After these plants had matured, at least two inflorescences were collected from 10 putative hybrids from each of the six different PMN crosses. All of the florets on these inflorescences were counted, threshed on a rubbing board, and cleaned through an air column of a South Dakota Seed Blower (E. L. Erickson Products, Brookings, S. D.) to determine seed set.
2.3. Flow Cytometry
Flow cytometry was used to determine the amount of DNA in the nuclei of the putative PMN hybrids, a pearl millet accession (PI 508273), and Merkeron napiergrass to predict their ploidy levels. Young leaves were collected from 10 putative hybrids from each of the crosses and was a diploid with 14 chromosomes, and it was used as an internal standard. Pieces of leaf blades, about 1 cm2, from the plant to be analyzed and the internal standard
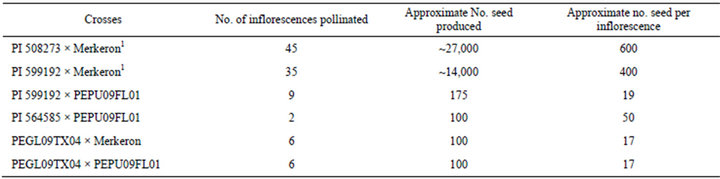
Table 2. Seed produced from the pearl millet by napiergrass pollinations.
1 based on no. inflorescences pollinated and average seed weights per 100 seed.
were placed into a 60 mm diameter plastic petri dish. Then 0.25 mL of Galbraith’s buffer was added to the dish and both leaf samples were finely chopped together with a razor blade. An additional 1.0 mL of Galbraith’s buffer was added to the macerated material, resuspended several times with a pipette, and the liquid was poured through a 30 μm filter into a 2.0 mL microtube. Fifty μL of propidium iodide was then added to each microtube and allowed to incubate for at least 15 minutes in a covered ice chest. The sample solutions were then analyzed for DNA content with a Partec CyFlow flow cytometer (Partec GmbH, Münster, Germany). The ploidy level of each putative PMN hybrid was determined by comparing the 2 C peak of the pearl millet standard with the 2 C peak of each putative hybrid. The same protocol was followed using napiergrass as the standard rather than pearl millet. A minimum of 3000 particles were analyzed for each sample. Each analysis was repeated three times.
2.4. EST-SSR Development
In the absence of publicly available sequence data for napiergrass, a total of 21,745 full-length complementary DNA (cDNA) sequences from apomictic buffelgrass [Pennisetum ciliare (L.) Link syn. Cenchrus ciliaris L.] pistils were downloaded from GenBank (National Center for Biotechnological Information http://www.ncbi.nlm.nih.gov/Genbank/index.html) [29]. Simple sequence repeats were identified and primer sequences were designed using the SSRLocator software package [30]. Selected SSRs contained at least 10 dinucleotide or five tri-, tetra-, or penta-nucleotide repeats. Primer design was based on the standards of 50% guaninecytosine content, minimum melting temperature of 50˚C, absence of secondary structure, length of 20 - 27 nucleotides, and amplified polymerase chain reaction (PCR) product range of 100 - 400 base pairs (bp) in length. The derived EST-SSRs were labeled “PCAR” (“PennisetumCiliare-Apomictic-Repeat”). A subset of 59 PCAR markers between 100 - 200 bp were selected and utilized in this study.
2.5. DNA Isolation
Genomic DNA was isolated using a modified rapid salt extraction protocol described by Aljanabi and Martinez [31]. Four-hundred µL of homogenizing buffer (0.4 M NaCl, 10 mM Tris-HCl, 2 mM EDTA, pH 8.0) and 100 mg of fresh leaf tissue were added to 1.7 mL microtubes. The plant tissue was pulverized for 1 to 2 minutes or until adequately pulped. Forty µL of 20% sodium dodecyl sulfate and 8 μL of 20 mg/mL proteinase K were added and vortexed for 5 seconds. Following incubation in a water bath at 65˚C for a minimum of 1 hour, 300 μL of NaCl saturated H2O was added and the samples were vortexed for 30 seconds. Samples were centrifuged at 12,000 rpm for 10 minutes, the supernatant was transferred into new tubes, the samples were centrifuged again at 12,000 rpm for 20 minutes, and supernatant was transferred into new tubes without disturbing the remaining pellets. Following the addition of 800 μL of cold isopropanol and 20 gentle inversions by hand, the samples were incubated at −20˚C for 1 hour. Samples were centrifuged at 10,000 rpm for 5 minutes, and the supernatant was removed. Then 500 μL of cold 70% ethanol was added to each tube. The samples were centrifuged at 10,000 rpm for 5 minutes, and the supernatant was removed. Microtubes containing DNA were inverted until dry, and the DNA was re-suspended in 100 μL of sterile deionized H2O. Re-suspended DNA was then quantified with a spectrophotometer (Eppendorf, Hamburg, Germany) prior to its use in PCR. After quantification, each sample was diluted to a final concentration of 50 ng μL−1.
2.6. Bulked Segregant Analysis
PCAR markers were used to survey the parental pearl millet and napiergrass genotypes. Equal amounts of DNA from each pearl millet genotype were combined into a bulk of maternal parents utilized in the PMN hybridizations. The maternal bulk was then surveyed in comparison to each of the two paternal napiergrass parents (Merkeron and PEPU09FL01). Polymerase chain reactions were performed in a total volume of 20 μL using 11.8 μL PCR H2O, 1 μL of 50 ng μL−1 DNA, 2 μL of 1X Promega MgCl2-free PCR buffer, 2 μL of 2.5 mM MgCl2, 1 μL of Taq polymerase. The reactions were conducted in 96-well plates and temperature cycling was carried out using a PTC-220 Dyad Thermal Cycler (MJ Research Inc., Waltham, MA). The PCR began with an initial denaturation at 95˚C for 3 minutes; followed by 40 touchdown decrement cycles at 95˚C for 25 seconds, 55˚C for 25 seconds, and 70˚C for 45 seconds; and concluded with an elongation stage of 72˚C for 10 minutes. The final hold was at 4˚C indefinitely. Amplification of the PCR products was completed using polyacrylamide gel electrophoresis (PAGE) on a large MEGA-GEL (C. B. S. Scientific, Del Mar, CA) high-throughput unit and nondenaturing gels with final concentrations of 40 mL acrylamide, 10 mL 10X TBE (tris-borate-EDTA) buffer, 1.4 mL ammonium persulfate, and 80 μL TEMED (Tetramethylethylenediamine) as described by Wang et al. [32]. The polyacrylamide gels were stained with ethidium bromide for 40 minutes prior to loading the DNA into the wells. Prior to loading the wells, 2 μL of gel loading buffer (35 mL of 50% glycerol, 2.5 mL of 10X TBE, 2 mL of 0.5M EDTA, 0.5 mL of 20% SDS, 10 mL of de-ionized H2O, and 0.05 g of bromophenol blue) was added to the 96 well plate and centrifuged up to a maximum of 340 rpm to thoroughly incorporate the loading buffer. After the PCR product and 1.5 μL of 50 bp ladder were loaded onto the gel rig, electrophoresis was carried out on a 2 hour run at a maximum amperage and wattage of 350 mA and 400 W, respectively. Since the desired bp length of the surveyed SSRs were 100 - 200, a run time of 2 hours was ideal. The identification of alleles that are 200 bp or larger requires 3 hours or more. Once the gels completed the electrophoresis process, they were photographed using UV light to illuminate the allele bands. The brightness, contrast, and white levels of the photographs were manipulated using Adobe Photoshop® and scored for the presence or absence of allele bands according to the procedure set forth by Rodriguez et al. [33].
3. Results and Discussion
3.1. Pearl Millet × Napiergrass Hybridization
The four pearl millet accessions were photoperiod insensitive with the number of days to flowering ranging from 50 to 120 days. Both napiergrass genotypes varied for photoperiod sensitivity, but neither initiated floral development early enough during the growing season to allow for field-based seed production. All pearl millet accessions produced viable seed when pollinated by napiergrass (Table 2). The average number of seed produced on each inflorescence was variable (Table 2). The identification and selection of pearl millet accessions with high seed set and napiergrass accessions with viable pollen are essential in developing a commercial PMN seed production system. Separation of putative hybrid seed into different size classes using sieves demonstrated that a wide range of seed sizes was produced. The average weight per seed for the four size classes were based on duplicated counts of 100 seed. For the progeny from the PI 508273 × Merkeron crosses, the weights were 7.36 mg for the 1.95 mm seed; 3.88 mg for the 1.81 mm seed; 3.02 mg for the 1.69 mm seed; and 2.14 mg for remaining class in this hybrid. For the progeny from the PI 599192 × Merkeron crosses, the weights were 7.78 mg for the 1.95 mm seed; 4.35 mg for the 1.81 mm seed; 3.65 mg for the 1.69 mm seed; and 2.36 mg for the 1.49 mm seed, the 1.49 mm seed. Germination was 100% for the 1.95 mm and 1.81 mm size classes and 90% for the 1.69 mm and 1.49 mm classes in this hybrid. Wrinkled seed coats were evident in the smallest seed class (1.49 mm) but did not affect germination. The size and weight differences in the PMN hybrid seeds indicate the need for further research to develop seeded PMN systems. Larger seed facilitate ease of planting, seedling emergence through crusted soil surfaces, increased ability to overcome weed pressure, and provide stronger seedlings in general to overcome these emergence stresses.
Phenotypic Traits and Seed Set
The plants that were produced from the above mentioned seed were regularly observed in the field. Some appeared similar to pearl millet but most more closely resembled napiergrass. Because of this, none of the offspring could be definitively classified as interspecific hybrids based solely on phenotypic traits.
The total number of flowering plants in each progeny row was used to determine fertility under open-pollinated conditions, and a minimum of two inflorescences were collected from each plant. The inflorescences were allowed to mature in the field for 4 weeks following pollination and were collected prior to seed shattering. Each inflorescence was used to determine the total number of florets produced and potential number of seed produced. After the florets were threshed and cleaned, no seed were recovered. This indicates that all the plants evaluated were sterile (Table 3). These plants are presumably interspecific hybrids because other researchers have reported that PMN hybrids are completely sterile [8,34-37]. Lack of seed production in F1 hybrids is one method whereby hybrids can be identified, but it is very timeconsuming because the plants cannot be evaluated until they have grown to This sterility is likely due to the differences in the ploidy levels and genomic compositions of the parents, which result in irregular meiosis in the F1 hybrids. Synteny between diploid (2n = 2x = 14; AA) pearl millet’s genome and one of allotetraploid (2n = 4x = 28; A’A’ BB) napiergrass’ sub genomes underlie this phenomenon. During metaphase I, members of the A genome of pearl millet tend to associate with members of the A’ genome from napiergrass to form as many as seven bivalents, and the chromosomes of the B genome from napiergrass are usually present as seven univalent [23]. Some of the univalents are subsequently not incorporated into the gametes because they lag behind the dividing chromosomes during anaphase I and II, resulting in sterility [36,38].
Flow Cytometry
When leaf tissue of different potential PMN hybrids and pearl millet was analyzed together, the 2C peaks of both plants overlapped and a difference in their DNA contents (data not shown) could not be detected. This was also the case when tissue of the PMN hybrids was analyzed with napiergrass tissue and when both pearl millet and napiergrass tissue were analyzed together. This indicates pearl millet and napiergrass have similar DNA contents and because of this, the 2C peaks of the putative PMN hybrids could not be separated from those of either parent. Consequently, it was impossible to determine if the putative hybrids were derived from self-fertilization of
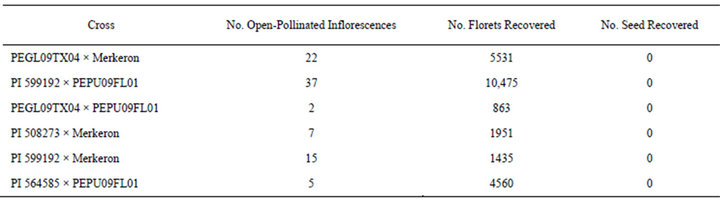
Table 3. Seed production from open-pollinated PMN hybrids.
pearl millet or were actually interspecific hybrids. Martel et al. [38] measured the DNA content of pearl millet and napiergrass and reported the 2C DNA content of pearl millet and napiergrass was 4.71 and 4.59 picograms, respectively. This explains why neither species could be separated from the putative PMN hybrids using flow cytometry. One would expect napiergrass to have approximately twice the DNA content as pearl millet and the hybrids would be intermediate between the two parental species because pearl millet is a diploid with 14 chromosomes and napiergrass is a tetraploid with 28 chromosomes. However, the pearl millet chromosomes are morphologically larger than the napiergrass chromosomes [38]. The mean chromosome length of pearl millet is 4.02 µm and 2.00 µm for napiergrass [38,39]. There is an insignificant difference for the mitotic total chromosome length in pearl millet and napiergrass which is 28.36 μm ± 3.92 and 28.09 μm ± 1.69, respectively [39]. This explains why the DNA quantities of the two species are similar even though napiergrass has twice as many chromosomes.
Hybrid Verification Using EST-SSRs
The use of molecular markers is essential in a successful wide-hybridization breeding program, and they are becoming more accessible and frequently used by breeders for the characterization of genetic diversity as well as population structure analysis in the corresponding germplasm of interest. Molecular markers can also be of value to distinguish heterotic groups from one another in a breeding population and to identify polymorphic regions within a heterotic group [40]. Markers that possess both gene based synteny across species and high polymerphism indexes, such as EST-SSRs, provide a reliable tool for identifying hybrids and analyzing genetic diversity using species-specific markers that are abundant in naturally heterogeneous species such as pearl millet and napiergrass [27]. However, the genomic resources readily available for use in pearl millet and napiergrass are limited. This drives the need to develop strategies for marker-assisted breeding in pearl millet, napiergrass, and their hybrids in combination with more recent interest for their use as biofuel feedstocks. To develop markers to use for PMN hybrid verification, 55 of the 59 EST-SSRs that were surveyed via bulked segregant analysis were successfully amplified using PCR. Numerous polymerphisms were detected and are summarized in Table 4. Four conserved markers, specific to the paternal napiergrass genotypes and absent in the maternal pearl millet genotypes, were chosen for marker-assisted hybrid verification provided their presence in the paternal parent (Table 5). Table 6 shows the overall summary of markers tested across parents in each respective cross and the number of progeny confirmed to be PMN hybrids. While beyond the objectives of this study, future utilization of co-dominant, heterozygous markers could further assist PMN breeding efforts by increasing the heterosis potential by selecting for greater levels of heterozygosity between parents. Future identification of pearl millet-specific PCAR markers would similarly facilitate markerassisted verification of Kinggrass (P. purpureum × P. glaucum), another seededyet-sterile biomass feedstock.
4. Conclusion
Our findings confirm previous reports that PMN hybridization is a feasible system and demonstrate its potential to overcome floral synchronization barriers by establishing greenhouse-crossing blocks during winter months. All PMN F1 hybrids evaluated in this study were completely sterile, and this agrees with previous reports. Breeding efforts to ensure selection of late flowering or non-flowering parental phenotypes therefore provide a strategy for developing high-biomass, “seeded-yetsterile” PMN cultivars. The putative hybrids could not be verified phenotypically or by their DNA content via flow cytometry. They could be identified by cytologically counting their chromosomes or by determining their seed fertility. However, both approaches are time consuming which delays selection of hybrids. The use of EST-SSRs is a more suitable method to confirm PMN hybrids. In
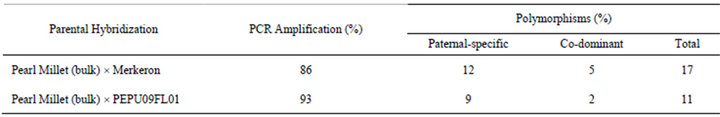
Table 4. Summary of bufflegrass EST-SSRs amplified across BSA of pearl millet and napiergrass.

Table 5. Primer sequences for selected “PCAR” EST-SSRs utilized for PMN hybrid verification.

Table 6. Survey of napiergrass specific EST-SSRs across PMN hybrids.
addition to delineating individual napiergrass parents and single crosses in this study, the conservation of several markers across multiple lines indicate conserved markers can be developed with specificity across all napiergrass germplasm in PMN hybridizations. Furthermore, recognition of genomic sequences that are deemed species-or cultivar-specific can be easily utilized in marker-assisted breeding approaches. Marker-assisted hybrid verification can also be utilized in the reciprocal cross of napiergrass × pearl millet as well as other wide hybrids involving either pearl millet or napiergrass as a parent. The EST-SSRs identified in this study further provide comparative genomic resources between napiergrass and major cereal crops.