Bacterial Biofilm in Water Bodies of Cherrapunjee: The Rainiest Place on Planet Earth ()
1. Introduction
Cherrapunjee (25˚17' - 25˚18'N and 91˚41'E - 91˚42'W), in India is the highest rainfall receiving area of the earth (average annual rainfall of 12,000 mm; Nov-Feb period being dry winter period with nominal or no rainfall and the rest eight months receive almost daily rains) where the soil is completely eroded due to deforestation and has only sparse grass cover. Consistent heavy rainfall in this wettest patch on earth may place an additional stress on the microbial communities. The water potential upshock associated with wetting of soil can kill a large fraction of the microbial community [1,2].
Biofilms are complex aggregation of microorganisms embedded in a self-produced matrix and adhering to inert or living surfaces [3]. They are characterized by structural heterogeneity, genetic diversity, complex community interactions, and an extracellular matrix of polymeric substances (EPS). This matrix protects the cells within it and facilitates communication among them through chemical and physical signals. EPS may vary in chemical and physical properties, but it is primarily composed of polysaccharides [4]. The major factors affecting the formation of biofilm on surfaces are electrochemical properties of the surface, nutrient availability and liquid flow [5]. The extent of microbial colonization appears to increase as the surface roughness increases due to the diminished surface area on rougher surfaces [6]. Maximum attachment depends upon higher surface free energy or wettability of surfaces. Surfaces with high surface free energies such as stainless steel and glass are more hydrophilic. These surfaces generally show greater bacterial attachment than hydrophobic surfaces such as Teflon, Buna-n rubber and fluorinated hydrocarbon. Solid surfaces which have been exposed in an aqueous medium become conditioned or coated with polymers from the medium. The chemical modification of surfaces affects the rate and extent of microbial attachment. The surface is converted to hydrophilic by cleaning with alkali or strong acid (4 M nitric acid) of stainless steel surfaces. Once the stainless steel is exposed to water, it is passivated by the formation of a chromium oxide layer [7]. Physico-chemical characteristics of aqueous medium such as pH, ionic strength, temperature, etc. may play an important role in the rate of microbial attachment to the surfaces. The bacterial attachment and biofilm formation in different aqueous systems are affected by season. This may be due to the temperature of water or other seasonally affected parameters. It is found that an increase in concentration of several cations such as sodium, calcium, lanthanum, ferric ions affects the attachment of P. fluorescens by reducing the repulsive forces between the cell and glass surfaces [6]. On the other hand, when nutrients are non-limiting in the liquid phase there is no need for the bacteria to attach themselves [5]. Stress situations like a depletion of nutrients makes sessile growth more favorable in flowing liquids [8].
We know of no reports discussing shifts in the biofilm bacterial community composition and activity as a result of long wet and short dry cycles. The proposed mechanism of succession in these three-dimensional communities is tolerance [9], which however, can assume different forms. Additionally, two nutrient groups were identified, namely tolerant, with low resource requirements (i.e. oligotrophic, mesotrophic and indifferent species) and sensitive, with high nutrient demands (i.e. meso-eutrophic to hyper-eutrophic forms) [10]. The majority of studies have explored the temporal trends of individual species in plants and much less so in microbial communities, arguing that interspecific interactions, especially competition, play a key role in community organization throughout succession in biofilm [11].
The present work was undertaken to analyze the diversity of bacteria involved in biofilm on stainless steel and glass substrata in water bodies of heaviest rainfall receiving patch on planet earth (map showing the site Cherrapunjee is depicted in Figure 1) and compare if the biofilm bacteria diversity in such heavy rainfall stress area had any shift in composition due to ecological uniqueness compared to other habitats.
2. Materials and Methods
2.1. Selection of Site
Water bodies located at Cherrapunjee (25˚17'N - 25˚18'N and 91˚41'E - 91˚42'E), India, the highest rainfall receiving place on planet earth were selected for study. Experiments were carried out during the months of JulyAugust which falls during the rainy season in this part of the country with the average temperature in the swelled water bodies due to incessant rain ranging from 12˚C - 15˚C.
2.2. Preparation and Cleaning of Surfaces for Biofilm Formation
The stainless steel coupon (2 × 5 cm) surfaces were rubbed with emery paper in order to remove edge corrosion tendency. The coupons were then cleaned with alcohol and air-dried. The glass slides were also rinsed and cleaned with alcohol, dried in air and kept in oven. The stainless steel coupons and the glass surfaces were kept at different depths in water bodies of Cherrapunjee separated by a distance of 15 cm for 15 days. Sampling was done in two sets, one on 7th day and the other on 15th day of incubation in the water bodies.
2.3. Isolation and Enumeration of Microorganisms
The different layers of stainless steel and glass slides were removed with sterile tongs and washed with phosphate buffered saline (PBS). The biofilm formed was removed with sterile swabs, which were transferred to

Figure 1. Map showing the studied Cherrapunjee area.
tubes containing 9 mL of 0.1% peptone water and spun in vortex for 2 min and was serially diluted upto 10–7. The dilutions from 10–4 to 10–7 were plated on Nutrient Agar (NA), Meat Agar (MA) and Brain Heart Infusion Agar (BHI) and were spread uniformly with a sterile spreader. Plates were incubated at 37˚C in an inverted position for 24 - 48 hr and the colony forming units (CFU) were recorded. Recovery ratios of bacterial cells (of both sets: on 7th day and 15th day) from the two substrates were calculated (so as to compare the number of bacteria between these substrata) as the highest CFU on stainless steel surface to that on glass surface. Further purifications of the isolates were done on respective plates and the isolates were maintained in NA slants at 5˚C. The isolates were also stored at –70˚C in Tryptone Soya Broth (TSB) containing 12.5% glycerol for future analysis [12].
2.4. Microscopic and Biochemical Characterization
The cell shape, size, arrangement and Gram character were observed for all the isolates under bright field microscope (Leica DM 5500, Germany). Preliminary identification and characterization was done by morphological and biochemical analysis following Bergey’s Manual of Determinative Bacteriology [13].
2.5. Scanning Electron Microscopy (SEM) Analysis
After the incubation period, one stainless steel coupon and one glass slide were taken (which showed white slimy layer), washed with PBS and immersed into a fixing solution (Karnovisk) at pH 7.2, and kept for 24 h. They were then washed three times for 10 min with sodium cacodylate buffer, and post-fixed in 1% osmium tetroxide for 1 h at room temperature. The substrates were washed three times with distilled water and dehydrated in a gradient series of acetone solutions (25%, 50%, 75%, 90%, 100%). The material reached its critical point using CO2 as the transitional fluid (Baltec CPD 030). Chips were attached to specimen stubs with conductive silver paint and the samples were gold-coated by sputtering for approximately 2 - 3 min in an evaporative coater (Baltec SCD 050). Finally, the stainless steel and glass surfaces were examined by scanning electron microscopy (JSM-6360, JEOL) [resolution: 3 nm in the secondary electron mode at working distance 8 mm; magnification 8× (WD 48 mm) to 300,000×; accelerating voltage 1 KV - 30 KV in 1 KV step].
2.6. Screening for Detection of Biofilm Formation
Evaluation and screening for detection of degree of biofilm formation by the isolates were carried out on Congo Red Agar (CRA) method and Tube Method (TM) [14,15].
2.7. Genomic DNA Isolation, 16S rRNA Gene Amplification and Sequencing
Six isolates were selected based on their varying colony morphology, biochemical characteristics and degree of biofilm formation. Genomic DNA isolation was done by HipurTM Bacterial Genomic DNA Purification Spin Kit (HiMedia, India) and the DNA bands were visualized in 0.8% agarose gel stained with ethidium bromide. Genomic DNA content and purity were estimated using a NanoVue Plus Spectrophotometer (GE Healthcare’s Life Sciences, Sweden). Bacterial 16S rDNA was amplified using the universal bacterial 16S rDNA primers, 27F [5’- AGA GTT TGA TCC TGG CTC AG - 3]’ and 1541R [5’- AAG GAG GTG ATC CAG CCG CA - 3’] [16] under the following conditions in Gene AMP PCR system 9700 (Applied Biosystems, California,USA): initial denaturation for 5 min at 94˚C, followed by 35 cycles consisting of denaturation at 94˚C for 1 min, annealing at 55˚C for 1 min, elongation at 72˚C for 2 min and then cycling was completed by a final elongation step for 5 min at 72˚C. A control tube containing sterile water instead of DNA solution was used as a negative control. PCR products were analyzed by electrophoresis in 1.5% (w/v) agarose gel in 1xTAE buffer with ethidium bromide (0.5 µg/mL). PCR products were purified using QIAquick Gel Extraction Kit (Qiagen, Germany). Sequencing reactions of the 16S rDNA fragments were performed with the Big Dye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, USA) using the above forward and reverse primers.
2.8. Phylogenetic Analysis
The 16S rRNA gene sequence of the isolates and their closest match were retrieved from EzTaxon server [http://www.eztaxon.org/] [17] and aligned using Clustal W with MEGA software version 4.1 [18] with Arcanobacterium pyogenes as the outgroup organism. Neighborjoining method [19] was employed to construct the phylogenetic tree with 1000 bootstrap replications to assess nodal support in the tree.
2.9. Study of Abiotic Stress Tolerance by the Isolates
Temperature tolerance: The isolates were inoculated in Luria-Bertani (LB) broth and incubated in 3 incubators maintained at different temperatures (5˚C, 30˚C, 55˚C). Salt tolerance: 3 sets of LB broth for each isolate were prepared with concentrations 0%, 12%, 24% of NaCl and were inoculated with the respective isolate. For pH tolerance, similar experiment was carried out with LB broth maintained at pH values of 3, 7 and 11 followed by simultaneous inoculation. All the above set of LB tubes were kept under shaking conditions for 24 hours. The temperature of incubation for the experiments on salt and pH tolerance was maintained at 37˚C. The growth was determined by optical density of the bacterial samples at 600 nm using spectrophotometer (CECIL Aquarius, CE 7200, UK) for all the above cases.
2.10. Extraction and Chemical Estimation of EPS
The bacterial cell suspension was centrifuged at 16,000 × g, 4˚C for 20 min [20]. The supernatants were filtered through a 0.22 μm membrane and the filtrate was used as the EPS sample [21]. Polysaccharide content in EPS was determined by the phenol sulphuric acid method [22], with glucose as the standard. The protein content of EPS was determined by the Bradford method [23] with bovine serum albumin as the standard.
3. Results
3.1. Enumeration and Characterization of Isolates
The diversity of bacterial colonies was higher in stainless steel surface than in glass surface incubated in the water bodies. CFU calculated on stainless steel surface ranged from 5.4 × 104 - 9.5 × 104 whereas too few colonies were encountered from the glass surfaces (Table 1). The recovery ratio of bacterial cells on stainless steel surface to glass surface was 1.7:1. CFU count was higher when sampling was done on the 15th day than on 7th day, but more morphologically diverse and distinct colonies were obtained on the 7th day. The isolates obtained from the two sampling days in various media were morphologically diverse (Table 2). A total of eleven distinct bacterial isolates were selected for study from stainless steel surfaces and one from glass surface. Preliminary screen-
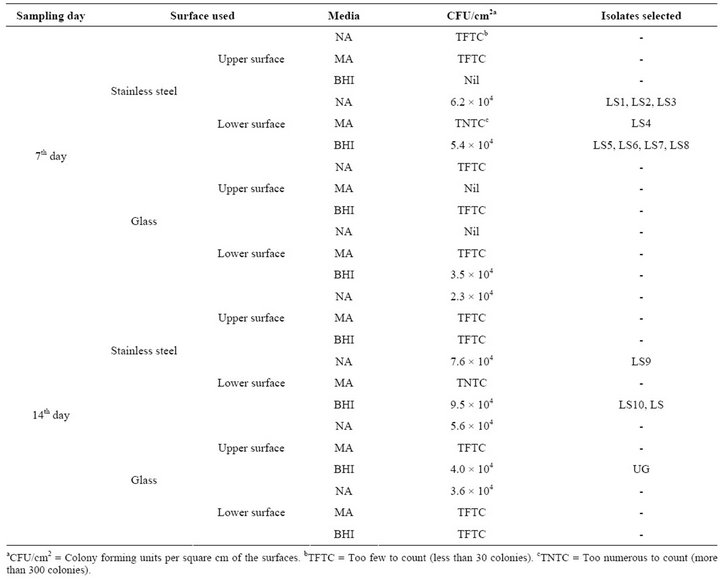
Table 1. Sampling parameters, culture media used and CFU counts of the isolates.
ing and morphological identifications showed two Gram positive and ten Gram negative bacteria, mostly rod shaped cells. There was variation in biochemical properties (Table 3) and growth pattern of the bacterial isolates in broth cultures (Table 4). Biochemical tests for the isolates showed close resemblance to the genera Bacillus, Proteus, Serratia, Pseudomonas, Chromobacterium and Achromobacter. The predominant bacteria among the population were Proteus and Pseudomonas.
3.2. SEM Analysis
We base our interpretation on the combination of the SEM photomicrographs that show the adherence of microbial filaments to the stainless steel (Figure 2(a)) and glass (Figure 2(b)). The electro micrographs obtained indicate a fairly thick biofilm formation.
3.3. Screening for Detection of Biofilm Formation
Analysis carried out by CRA and TM assay revealed weak, moderate and strong biofilms (Table 4). The CRA assay showed good correlation with the TM assay for moderate biofilm forming isolates and four isolates were picked up as strong and three were weak biofilm producers. In the TM assay, only two isolates displayed high degree of biofilm formation.

Table 2. Morphological characteristics observed among the isolates.

Table 3. Biochemical characterization of the isolates and tentative match.
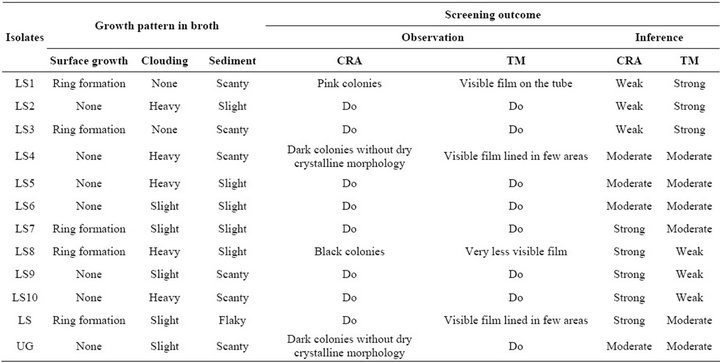
Table 4. Growth pattern in broth cultures and screening outcome for the isolates.
 (a) (b)
(a) (b)
Figure 2. Electromicrographs of bacterial cells adhered onto (a) stainless steel surface and (b) glass surface visualized by scanning electron microscopy [arrows indicate the biofilm layer that can be observed in each image].
3.4. PCR Amplification of 16S rDNA and Phylogenetic Analysis
Two distinct clusters were observed in the phylogenetic tree, one including all Gram negative bacteria and the other containing Gram positive Bacillus sp. along with the outgroup organism (Figure 3).
3.5. Nucleotide Sequence Accession Numbers
Nucleotide sequences were deposited in GenBank under the accession numbers: JN695728, JN566135, JN566136, JN566137, JN653472 and JN653473 (Table 5).
3.6. Abiotic Stress Tolerance and EPS Content
Abiotic stress tolerance at various temperatures showed maximum growth of all isolates at 30˚C, while extremely low growth was obtained at 5˚C and 55˚C (Figure 4). pH 7.0 was observed as the optimum while growth rate was low at pH 3 and 11. An exception to the case is with the
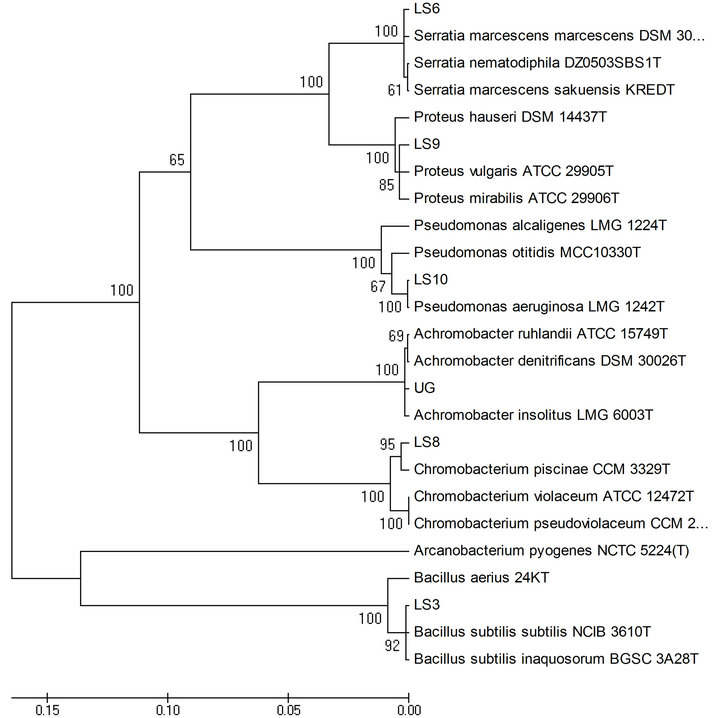
Figure 3. Phylogenetic tree constructed for the isolates using neighbor joining method.
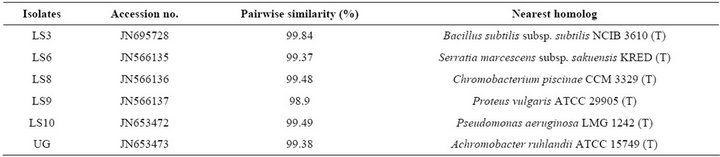
Table 5. Molecular analysis of selected six isolates and accession numbers.
isolate LS3 which grew well in an alkaline pH 11 than at pH 7.0, but growth in acidic pH was very low (Figure 5). The growth of bacteria was affected by salt concentration (Figure 6). There was significant difference in protein and carbohydrate content in the EPS extracted from different isolates (Figure 7).
4. Discussion
Due to its important role in the environment, industry and medicine, the understanding of mechanisms of biofilm formation has become the focus of biofilm research. A wide range of microorganisms are capable of forming biofilms, and as a result, biofilms exist in a variety of environment [8]. This study provides basic information about the diversity of biofilm bacteria associated with the heaviest rainfall receiving water bodies of Cherrapunjee. In general, it is assumed that glass and stainless steel are hydrophilic materials while rubber and plastic are hydrophobic

Figure 4. Growth pattern of isolates at different temperatures.

Figure 5. Growth pattern of isolates at different pH.

Figure 6. Growth pattern of isolates at different salt concentrations.

Figure 7. Protein and carbohydrate content in EPS extracted from the isolates.
materials [6,24]. It has been previously shown that micro-organisms, including Salmonella spp. and L. monocytogenes strains, adhere in higher numbers to more hydrophobic materials [24]. Bacteria in an aquatic environment may be exposed to different levels of nutrients, depending upon the location of the water body [25]. On the other hand, it is well known that many factors influence biofilm formation, among them the composition of the medium is important [26-29]. Therefore, three different media were selected for this study. Our study revealed that irrespective of the duration of association of the two substrates with the water bodies, the attachment and colonization of biofilm bacteria was more on stainless steel than that of glass surface.
Among the various screening methods for detection of degree of biofilm formation, we obtained very little correlation among the two methods as the parameters of sensitivity, specificity and accuracy were very low, which was in agreement with observations of other investigators [14]. The in vitro stress tolerance showed no unusual tolerance behavior among the characterized biofilm bacteria.The nature of the diversity of biofilm bacteria in the present study confirms with the diversity reported in other conditions [30], indicating that heavy rainfall stress has no significant uniqueness and shift effect on biofilm composition.
5. Acknowledgements
Authors acknowledge the grant received from Department of Information Technology (Ministry of Communications & Information Technology), Govt. of India, and thank Sophisticated Analytical Instrument Facility (SAIF), NEHU, Shillong for providing the SEM services.
NOTES