Advances in Microbiology
Vol.3 No.1(2013), Article ID:29127,6 pages DOI:10.4236/aim.2013.31007
Metabolic Engineering of Thermoanaerobacterium thermosaccharolyticum for Increased n-Butanol Production
1Dartmouth College, Hanover, USA
2Mascoma Corporation, Lebanon, USA
Email: ashwini.bhandiwad.th@dartmouth.edu, anna.guseva@dartmouth.edu, *lee.r.lynd@dartmouth.edu
Received January 8, 2013; revised February 8, 2013; accepted March 5, 2013
Keywords: Biofuels; n-Butanol; Thermophile; Metabolic Engineering
ABSTRACT
Thermoanaerobacterium thermosaccharolyticum shows promise as a host for n-butanol production since it natively has the required genes involved in the n-butanol biosynthetic pathway. Overexpression of the natively occurring bcs operon containing the genes thl, hbd, crt, bcd, etfA, and etfB responsible for the formation of butyryl CoA increased the n-butanol production by 180% compared to the wild type from a n-butanol titer of 1.8 mM to 5.1 mM. The deletion of one of the six alcohol dehydrogenase genes confirmed that it was the primary gene responsible for ethanol and n-butanol production from acetyl CoA and butyryl CoA respectively.
1. Introduction
In recent years, cellulosic biofuels have gained much interest as an alternative to petroleum as transportation fuel. Besides ethanol, butanol is being considered as a potential next-generation biofuel. The four-carbon alcohol has been studied since 1920s as a product of clostridial fermentations [1]. The energy density and octane rating of n-butanol molecule is comparable to gasoline and can be blended with gasoline or used 100% as a fuel without engine modification. Additionally n-butanol is less corrosive and has a low enough vapor pressure that allows it to be transported via existing the infrastructure for petroleum [2]. There have been intensive ongoing efforts towards engineering a range of mesophilic organisms [3-6] including the native Clostridial n-butanol producers for increased yields and titers of n-butanol [7-11]. However, to date, engineering of thermophilic anaerobes for the production of n-butanol has not been reported. Thermophiles offer potential advantageous features compared to mesophiles for use in industrial processes [12].
T. thermosaccharolyticum (formerly called Clostridiumthermosaccharolyticum) was originally isolated in 1930s as spoilage from canned foods [13,14]. This microbe produces n-butanol at a yield of 0.03 mol/mol of glucose equivalent under non-pH controlled conditions besides producing lactic acid, acetic acid, butyric acid, and ethanol [15]. T. thermosaccharolyticumhas been widely studied in the context of conversion of cellulosic biomass to ethanol. However, consistent n-butanol formation has not been reported [16-18].
The genome of T. thermosaccharolyticum DSM 571 became publicly available in 2010 [19]. Exploration of the genome indicated that the genes responsible for butyryl CoA production occur asabcs operon comprising of the genesenoyl-CoA hydratase (crt) (Tthe_1661), acylCoA dehydrogenase (bcd) (Tthe_1660), electron transfer flavoprotein alpha/beta subunits(etfA/B) (Tthe_1658, Tthe_1659), 3-hydroxyacyl-CoA dehydrogenase (hbd) (Tthe_1657), and acetyl-CoA acetyltransferase (thl) (Tthe_1656). Deletion of competing pathways can increase the carbon flux towards n-butanol. Putative genes responsible for lactic acid production—L-lactate dehydrogenase (ldh) (Tthe_2412), and acetic acid production —phosphate acetyltransferase (pta) (Tthe_1502) and acetate kinase (ack) (Tthe_1501) have been identified as targets for deletion. There are six alcohol dehydrogenase genes identified on the genome, however only one (Tthe_2646) has sequence similarity to the conserved domains of the bi-functional aldehyde-alcohol dehydrogenase thought to be used for catabolic reduction of acetyl CoA to ethanol in Thermoanaerobacterium sp. and Thermoanaerobacter sp. [20]. The gene Tthe_2646 (adhE) is also assumed to be responsible for the butyryl CoA reduction to n-butanol. To confirm the functionality of this gene, it was targeted for deletion. In clostridia, the genes responsible for the formation of butyrate are phosphotransbutyrylase (ptb) and butyrate kinase (bk) [1,21-23]. However, similar genes do not appear to be present on the genome of T. thermosaccharolyticum DSM 571.
T. thermosaccharolyticumis naturally competent [24], utilizes hemicellulose sugars, and has temperature and pH optima compatible with cellulolytic thermophiles such as C. thermocellum. This study was undertaken with the objective of using metabolic engineering to increase n-butanol production.
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
T. thermosaccharolyticumstrain 571 was obtained from Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ, Germany). The bacterial strains and plasmids used are listed in Table 1. For transformation experiments all strains were grown in TSC1 medium [25] at 55˚C, containing per liter—5 g of cellobiose, 1.85 g of (NH4)2SO4, 0.05 g of FeSO4·7H2O, 1.0 g of KH2PO4, 1.0 g of MgSO4, 0.1 g of CaCl2·2H2O, 2 g of Na3C6H5O7·2H2O, 5 g of yeast extract, 0.002 g of resazurin, 0.5 g of L-cysteine-HCl; and for solid media 12 g of agarose used. The pH was adjusted to 6.7 for selection on kanamycin (200 mg/mL).
2.2. Construction of Recombinant Strains of T. thermosaccharolyticum
The native genes of T. thermosaccharolyticum were over
Table 1. Strains and plasmids used.

expressed on a replicative plasmid pBu-bcs. The expression of the genes crt, bcd, etfB, etfA, hbd and thl was mediated by the glyceraldehyde-3-phosphate dehydrogenase (gapDH) promoter from C. thermocellum. Non-replicative plasmids pTTLAKO, pTTAAKO and pTTADHEKO were constructed in order to disrupt the lactate, acetate and alcohol production pathways respectively. All the plasmids were constructed using Gibson Assembly using E. coli cells [26] in which each fragment in the construction that was amplified using PCR had an overlap region of 35 bp with the adjacent fragments. The flanking upstream and downstream regions of homology contained a kanamycin resistance gene [27] that replaced the genes ldh; pta and ack; and adhE. Transformation was carried out in an anaerobic chamber (Coy laboratories, Grass Lake MI) by inoculating 10 mL of TSC1 medium with 1 mL of frozen parent strain stocks. 1 mL of the cultures were added to a tube containing approximately 500 ng of plasmid DNA. The cultures were incubated for 15 - 18 hours at 55˚C and dilutions were suspended in molten TSC1 agar at pH 6.7 and allowed to solidify before incubation at 55˚C [24]. Selection of the positive clones was done by growing on 200 mg/mL kanamycin supplemented media. Five such positive clones were selected for further analysis.
2.3. Transcript Analysis
Cells were harvested in mid-log phase and stabilized with RNA protect. The RNA extraction was performed using RNeasy Kit (Qiagen, Valencia CA) along with the on-column DNAse treatment. The qPCR was conducted using the iScript One-Step RT-PCR Kit with SYBR Green (Bio-rad, Hercules CA) Top of Form. Primers were designed for targeting 150 - 200 bp internal regions of the genes of interest.
2.4. End Product Determination
The fermentation end products were measured by highpressure liquid chromatography with an Aminex HPX- 87 H column (Bio-rad Laboratories, Hercules CA). The mobile phase consisted of 5 mM sulfuric acid at a flow rate of 0.6 mL/min. The detection of the metabolites was by refractive index using a refractometer (Waters 410).
2.5. Growth Comparisons
The growth comparisons between the strains constructed was carried out using a plate reader (BioTek, Winooski VT) placed inside an anaerobic chamber. 200 mL TSC1 media was inoculated with 10 mL of an overnight grown culture. The cultures were allowed to grow at 55˚C until each strain reached stationary phase of growth. Each measurement was carried out in triplicate.
2.6. Statistical Analysis
A one-tailed statistical test was applied to examine the significance of end product formation results from the different T. thermosaccharolyticum strains constructed. A critical value of significance was considered a p-value less than 0.05. The Graph Pad software was used to analyze and calculate the p-values.
3. Results
3.1. Construction of T. thermosaccharolyticum Recombinant Strains
Transformation of T. thermosaccharolyticum strain DSM 571 with the replicative plasmid pBu-bcs containing the genes in the bcs operon driven by gapDH promoter (Figure 1) resulted successful transformants with an efficiency of 103 transformants/mg plasmid DNA. Five transformants were selected for verification of plasmid DNA and n-butanol production. One strain designated as TTbcs was selected for further characterization.
Transformation of T. thermosaccharolyticum strain DSM 571 with non-replicative plasmids pTTLAKO (Figure 2) and pTTADHEKO resulted in successful transformants with similar efficiency of 103 transformants/mg plasmid DNA. After verification of chromosomal integration at the desired locus, one strain each for

Figure 1. Construction of replicative plasmid pBu-bcs.
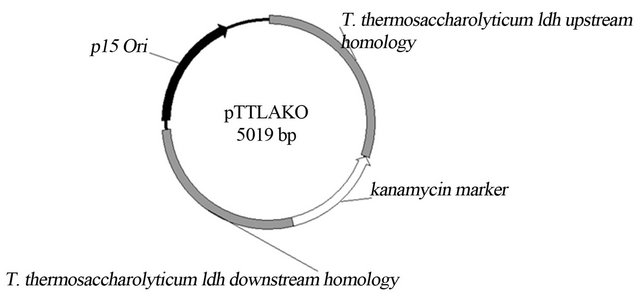
Figure 2. Non-replicative plasmid construction plasmid for lactate deletion (pTTAAKO and pTTADHEKO were constructed similarly).
ldh deletion and adhE deletion was selected and designnated Δldh and ΔadhE respectively. Multiple efforts to delete pta and ack were unsuccessful in achieving a ΔptaΔack strain.
3.2. Transcript Analysis
The transcription levels of each of the genes in strain TT-bcs—thl, hbd, crt, bcd, etfA and etfB were 3 - 4 times higher than the WT strain (Figure 3). Although adhE was not overexpressed in TT-bcs, the transcript levels were seen to be elevated almost 28 fold higher than the WT strain.
The quantitative reverse transcriptase PCR data confirmed that the ldh gene was not transcribed in strain Δldh. The transcript levels of genes thl, hbd, crt, bcd, etfA, etfB and adhE increased approximately 2-fold compared to the WT strain.
The qPCR data confirmed that adhE was not transcribed in strain ΔadhE. Interestingly, ldh transcript levels in strain ΔadhE were 32-fold higher than the WT strain.
3.3. End Product Analysis
n-Butanol production by strain TT-bcs increased by 180%, from 1.8 mM to 5.1 mM compared to the WT strain. Statistical analysis illustrated that this was a significant increase with a p-value of 0.003. A cetate production increased in strain TT-bcs by 20% from 24.8 mM to 30 mM compared to the WT strain (statistically significant with a p-value of 0.03). Further, butyric acid production decreased by 20% in TT-bcs compared to WT 34.8 mM to 27.8 mM (statistically significant with a p-value of 0.002) (Table 2).

Figure 3. Normalized expression levels of genes ldh, ack, thl, hbd, crt, bcd, etfB, etfA and adhE in four strains of T. thermosaccharolyticum relative to 16S expression WT (solid grey), Δldh (black dots), ΔadhE (solid black) and TT-bcs (black stripes). Data represents an average of triplicates with error bars.
In strain Δldh, lactate production was not detected confirming the functionality of the ldh gene. The end product spectrum shifted to allow an increase of 8% in butanol production, although statistically this was insignificant (p-value of 0.38).
Ethanol and n-butanol production was not detected in strain ΔadhE. This was accompanied by a 17-fold increase in lactic acid production compared to WT strain from 1.8 mM to 31.1 mM. Moreover, no acetate or butyrate production was detected in strain ΔadhE.
3.4. Growth Comparisons
The growth rate of the TT-bcs strain was comparable to the WT (Figure 4). However, the maximum cell density reached was compromised by approximately 10%. The specific growth rate of the Dldh strain (0.23 h−1) decreased by 20% compared to the wild type (0.29 h−1). The DadhE strain grew very poorly, with a specific growth rate of 0.033 h−1 compared to 0.29 h−1 for the wild type strain.
4. Discussion
In order to make T. thermosaccharolyticum aviable host for n-butanol production, multiple efforts need to be undertaken to channel the maximum possible carbon and electron flux through the n-butanol pathway. To understand this concept more thoroughly, we studied the effect of overexpressing the bcs operon containing the gene products for n-butanol production, as well as targeting acetate, lactate, and alcohol pathways for deletion.
Overexpression of the genes responsible for butyryl CoA formation not only led to elevated transcription levels of thl, hbd, crt, bcd, etfA, and etfB, but also increased transcription of adhE. Although the over expression of the bcs operon genes increased n-butanol production by 180% compared to WT levels, this translated to only 9% of the maximum theoretical yield.
The phenotype of strain ΔadhE confirmed that the targeted gene adhE out of the six identified alcohol dehydrogenase genes, is responsible for both ethanol and butanol production since neither of the alcohols (ethanol or n-butanol) were detected by HPLC in the DadhE strain. Deletion of adhE caused increased lactate production, to roughly 1600% WT levels. The carbon flow is diverted primarily to the lactic acid pathway.
However in this study, targeted gene deletions of the primary acetate pathway involving the genes pta and ack proved ineffective, and additional effort needs to be applied in order to effectively eliminate acetate production.
Although this study has demonstrated some improvements over the WT for n-butanol production, additional work is required to realize the application of T. thermosaccharolyticum as a feasible host for n-butanol production in an industrial process.
5. Acknowledgements
This research was supported by Mascoma Corporation, Lebanon, NH and a grant from the BioEnergy Science Center (BESC). We would like to thank Dr. Joe Shaw for providing strains and plasmids. We would also like to thank Elizabeth Mearls for critical review of the manuscript and Phil Thorne from Mascoma Corp. for contributing

Figure 4. Growth comparison between strains of T. thermosaccharolyticum WT (red), TT-bcs (green), Δldh (blue), ΔadhE (yellow). Data represents an average of triplicate experiments performed at 55˚C in a plate reader.
Table 2. End product profiles of T. thermosaccharolyticum strains.

Data represents batch fermentations in triplicates with 10 g initial cellobiose/l.
to the analytical methods.
REFERENCES
- D. T. Jones and D. R. Woods, “Acetone-Butanol Fermentation Revisited,” Microbiology Review, Vol. 50, No. 4, 1986, pp. 484-524.
- P. Durre, “Fermentative Butanol Production: Bulk Chemical and Biofuel,” Annals of the New York Academy of Sciences, Vol. 1125, 2008, pp. 353-362.
- S. Atsumi, A. F. Cann, M. R. Connor, C. R. Shen, K. M. Smith, M. P. Brynildsen, K. J. Chou, T. Hanai and J. C. Liao, “Metabolic Engineering of Escherichia coli for 1-Butanol Production,” Metabolic Engineering, Vol. 10, No. 6, 2008, pp. 305-311. doi:10.1016/j.ymben.2007.08.003
- M. Inui, M. Suda, S. Kimura, K. Yasuda, H. Suzuki, H. Toda, S. Yamamoto, S. Okino, N. Suzuki and H. Yukawa, “Expression of Clostridium Acetobutylicum Butanol Synthetic Genes in Escherichia coli,” Applied and Environmental Microbiology, Vol. 77, No. 6, 2008, pp. 1305- 1316. doi:10.1007/s00253-007-1257-5
- D. R. Nielsen, E. Leonard, S. H. Yoon, H. C. Tseng, C. Yuan and K. L. Prather, “Engineering Alternative Butanol Production Platforms in Heterologous Bacteria,” Metabolic Engineering, Vol. 11, No. 4-5, 2009, pp. 262-273. doi:10.1016/j.ymben.2009.05.003
- B. B. Bond-Watts, R. J. Bellerose and M. C. Y. Chang, “Enzyme Mechanism as a Kinetic Control Element for Designing Synthetic Biofuel Pathways,” Nature Chemical Biology, Vol. 7, No. 4, 2011, pp. 222-227. doi:10.1038/nchembio.537
- L. D. Mermelstein, E. T. Papoutsakis, D. J. Petersen and G. N. Bennett, “Metabolic Engineering of Clostridium Acetobutylicum ATCC 824 for Increased Solvent Production by Enhancement of Acetone Formation Enzyme Activities Using a Synthetic Acetone Operon,” Biotechnology and Bioengineering, Vol. 42, No. 9, 1993, pp. 1053-1060. doi:10.1002/bit.260420906
- E. T. Papoutsakis, “Engineering Solventogenic Clostridia,” Current Opinion in Biotechnology, Vol. 19, No. 5, 2008, pp. 420-429. doi:10.1016/j.copbio.2008.08.003
- T. Lutke-Eversloh and H. Bahl, “Metabolic Engineering of Clostridium Acetobutylicum: Recent Advances to Improve Butanol Production,” Current Opinion in Biotechnology, Vol. 22, No. 5, 2011, pp. 634-647. doi:10.1016/j.copbio.2011.01.011
- R. Sillers, A. Chow, B. Tracy and E. T. Papoutsakis, “Metabolic Engineering of the Non-Sporulating, Non-Solventogenic Clostridium Acetobutylicum Strain M5 to Produce Butanol without Acetone Demonstrate the Robustness of the Acid-Formation Pathways and the Importance of the Electron Balance,” Metabolic Engineering, Vol. 10, No. 6, 2008, pp. 321-332. doi:10.1016/j.ymben.2008.07.005
- R. Sillers, M. A. Al-Hinai and E. T. Papoutsakis, “Aldehyde-Alcohol Dehydrogenase and/or Thiolase Over-Expression Coupled with CoA Transferase Down Regulation Lead to Higher Alcohol Titers and Selectivity in Clostridium Acetobutylicum Fermentations,” Biotechnology and Bioengineering, Vol. 102, No. 1, 2009, pp. 38-49. doi:10.1002/bit.22058
- J. G. Zeikus, “Thermophilic Bacteria—Ecology, Physio logy and Technology,” Enzyme and Microbial Technology, Vol. 1, No. 4, 1979, pp. 243-252. doi:10.1016/0141-0229(79)90043-7
- L. S. McClung, “Studies on Anaerobic Bacteria: III. Historical Review and Technique of Culture of Certain Thermophilic Anaerobes,” Journal of Bacteriology, Vol. 29, No. 2, 1935, pp. 173-187.
- L. S. McClung, “Studies on Anaerobic Bacteria: IV. Taxonomy of Cultures of a Thermophilic Species Causing ‘Swells’ of Canned Foods,” Journal of Bacteriology, Vol. 29, No. 2, 1935, pp. 189-203.
- D. Freierschroder, J. Wiegel and G. Gottschalk, “Butanol Formation by Clostridium-Thermosaccharolyticum at Neutral Ph,” Biotechnology Letters, Vol. 11, No. 11, 1989, pp. 831-836. doi:10.1007/BF01026107
- J. N. Saddler and M. K. H. Chan, “Conversion of Pretreated Lignocellulosic Substrates to Ethanol by Clostridium Thermocellum in Monoand Co-Culture with Clostridium Thermosaccharolyticum and Clostridium Thermohydrosulphuricum,” Canadian Journal of Microbiology, Vol. 30, No. 2, 1984, pp. 212-220. doi:10.1139/m84-032
- A. L. Demain, M. Newcomb and J. H. Wu, “Cellulase, Clostridia, and Ethanol,” Microbiology and Molecular Biology Reviews, Vol. 69, No. 1, 2005, pp. 124-154. doi:10.1128/MMBR.69.1.124-154.2005
- N. O. Sjolander, “Studies on Anaerobic Bacteria: XII. The Fermentation Products of Clostridium Thermosaccharolyticum,” Journal of Bacteriology, Vol. 34, No. 4, 1937, pp. 419-428.
- C. L. Hemme, H. Mouttaki, Y. J. Lee, G. Zhang, L. Goodwin, S. Lucas, A. Copeland, A. Lapidus, T. Glavina del Rio, H. Tice, E. Saunders, T. Brettin, J. C. Detter, C. S. Han, S. Pitluck, M. L. Land, L. J. Hauser, N. Kyrpides, N. Mikhailova, Z. He, L. Wu, J. D. Van Nostrand, B. Henrissat, Q. He, P. A. Lawson, R. S. Tanner, L. R. Lynd, J. Wiegel, M. W. Fields, A. P. Arkin, C. W. Schadt, B. S. Stevenson, M. J. McInerney, Y. Yang, H. Dong, D. Xing, N. Ren, A. Wang, R. L. Huhnke, J. R. Mielenz, S. Y. Ding, M. E. Himmel, S. Taghavi, D. van der Lelie, E. M. Rubin and J. Zhou, “Sequencing of Multiple Clostridial Genomes Related to Biomass Conversion and Biofuel Production,” Journal of Bacteriology, Vol. 192, No. 24, 2010, pp. 6494-6496. doi:10.1128/JB.01064-10
- S. Yao and M. J. Mikkelsen, “Identification and Over-Expression of a Bifunctional Aldehyde/Alcohol Dehydrogenase Responsible for Ethanol Production in Thermoanaerobacter Mathranii,” Journal of Molecular Microbiology and Biotechnology, Vol. 19, No. 3, 2010, pp. 123-133. doi:10.1159/000321498
- Y. Zhang, M. Yu and S. T. Yang, “Effects of ptb KnockOut on Butyric Acid Fermentation by Clostridium Tyrobutyricum,” Biotechnology Progress, Vol. 28, No. 1, 2012, pp. 52-59. doi:10.1002/btpr.730
- X. Liu, Y. Zhu and S. T. Yang, “Construction and Characterization of Ack Deleted Mutant of Clostridium Tyrobutyricum for Enhanced Butyric Acid and Hydrogen Production,” Biotechnology Progress, Vol. 22, No. 5, 2006, pp. 1265-1275. doi:10.1021/bp060082g
- C. Zhang, H. Yang, F. Yang and Y. Ma, “Current Progress on Butyric Acid Production by Fermentation,” Current Microbiology, Vol. 59, No. 6, 2009, pp. 656-663. doi:10.1007/s00284-009-9491-y
- A. J. Shaw, D. A. Hogsett and L. R. Lynd, “Natural Competence in Thermoanaerobacter and Thermoanaerobacterium Species,” Applied and Environmental Microbiology, Vol. 76, No. 14, 2010, pp. 4713-4719. doi:10.1128/AEM.00402-10
- A. J. Shaw, S. F. Covalla, D. A. Hogsett and C. D. Herring, “Marker Removal System for Thermoanaerobacterium Saccharolyticum and Development of a Markerless Ethanologen,” Applied and Environmental Microbiology, Vol. 77, No. 7, 2011, pp. 2534-2536. doi:10.1128/AEM.01731-10
- D. G. Gibson, L. Young, R. Y. Chuang, J. C. Venter, C. A. Hutchison and H. O. Smith, “Enzymatic Assembly of DNA Molecules Up to Several Hundred Kilobases,” Nature Methods, Vol. 6, No. 5, 2009, pp. 343-U41. doi:10.1038/nmeth.1318
- V. Mai, W. W. Lorenz and J. Wiegel, “Transformation of Thermoanaerobacterium sp. Strain JW/SL-YS485 with Plasmid pIKM1 Conferring Kanamycin Resistance,” FEMS Microbiology Letters, Vol. 148, No. 2, 1997, pp. 163-167. doi:10.1111/j.1574-6968.1997.tb10283.x
NOTES
*Corresponding author.

