American Journal of Plant Sciences
Vol.07 No.03(2016), Article ID:64405,8 pages
10.4236/ajps.2016.73039
Growth and Physiological Attributes of Tomato (Lycopersicon esculentum Mill.) Genotypes as Affected by NaCl Stress
M. Nasir Khan
Department of Biology, Faculty of Science, University of Tabuk, Tabuk, Saudi Arabia

Copyright © 2016 by author and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 29 January 2016; accepted 6 March 2016; published 10 March 2016
ABSTRACT
High level of salinity adversely affects growth, productivity as well as quality of vegetable crops by reducing photosynthetic capacity, enzyme activities, and by enhanced production of reactive oxygen species (ROS). These ROS cause peroxidation of membrane lipids leading to disintegration of cell membrane with the leakage of electrolytes. All these detrimental effects ultimately contribute to the crop with reduced yield and low nutritive value. The present investigation was carried out to test salt tolerance capacity of ten genotypes of tomato on the basis of growth, physiological and biochemical characteristics. The results show that all the ten tested genotypes of tomato responded differently to 200 mM NaCl stress. Regarding growth parameters, BL-1076 gave higher values, while Queen gave the lowest values for most of the parameters (plant height, number of leaves, leaf area, shoot and root fresh weight and dry weight). Regarding physiological and biochemical parameters, BL-1076 gave higher values for the activities of carbonic anhydrase and nitrate reductase, leaf chlorophyll content, relative water content, and activities of antioxidant enzymes (superoxide dismutase, peroxidase and catalase). However, genotype Queen gave the lowest values for these parameters. On contrary, the least membrane damage (TBARS content) was registered in BL- 1076, while the highest in Queen. Overall assessment of results leads to the conclusion that the genotype BL-1076 was found salt tolerant while Queen salt sensitive. The ten genotypes may be arranged on the basis of their salt tolerance capacity in decreasing order as: BL-1076 > Trust > Imperial > Tanshet star > PakmoreVF > L 26 > Plitz > Bonus F1 > Grace > Queen.
Keywords:
Antioxidant Enzymes, Genotypes, Growth, Stress, Tomato

1. Introduction
Tomato fruits are one of the important horticultural crops grown across the globe. Tomatoes are an excellent source of antioxidants, dietary fiber, minerals, and vitamins [1] -[3] and are one of the low-calorie vegetables; hold just 18 calories per 100 g. Total ORAC (Oxygen Radical Absorbance Capacity) or antioxidant power of tomato has been noted as high as 1341 µmol TE/100g [4] . In spite of being such an important commodity, production and quality of tomato does not meet the requirements, because low humidity and high temperature of Tabuk region jointly accelerate the rate of evaporation which leaves behind the accumulated salts. High concentration of NaCl, the dominant salt in the soil, poses osmotic stress and ionic stress, which reduces the ability of plants to take up water and minerals [5] [6] . Moreover, hyper accumulation of sodium (Na+) ion in the cytosol damages cell membrane leading to leakage of electrolytes and affects metabolic activities in cytosol [7] . The results of these effects lead to depressed physiological and biochemical processes. High level of salinity adversely affects growth, productivity as well as quality of vegetable crops by reducing photosynthetic capacity, enzyme activities by enhanced production of reactive oxygen species (ROS) which causes accumulation of malondialdehyde (MDA) content, which is one of the final products of peroxidation of membranes lipids [8] . Lipid peroxidation leads to disintegration of cell membrane with the leakage of electrolytes. All these detrimental effects ultimately contribute to the crop with reduced yield and low nutritive value. However, to scavenge ROS plants are equipped with a system of antioxidant enzymes such as superoxide dismutase (SOD), peroxidase (POX), and catalase (CAT). SOD converts superoxide radicals to H2O2, while CAT and POX dismutate H2O2 into water and oxygen. However, precise and timely activation of these antioxidant enzymes is vital so that plant cells could be protected before the onset of any oxidative damage. It is a well known fact that plants show significant variation to the environmental stimuli even at intra-specific level, thus timely and precise response to an adverse stimulus, like salt stress, will decide the fate of the crop plant. A plant will show improved tolerance to salinity if it senses the stress stimulus as early as possible and causes activation of its defence system. Therefore, to cope with the detrimental situation of salt stress and to avoid losses due to salinity, identification of salt sensitive and tolerant genotypes of tomato is of foremost importance. Considering the same in mind, the present investigation was carried out to select salt sensitive and salt tolerant genotypes of tomato on the basis of growth and physiological attributes.
2. Materials and Methods
2.1. Plant Culture
Surface sterilized healthy seeds of ten genotypes of tomato (Lycopersicon esculentum Mill.), namely BL-1076, Trust, Imperial, Tanshet star, PakmoreVF, L 26, Plitz, Bonus F1, Grace and Queen were sown in seedling trays. 30 days after sowing (DAS), seedlings were transplanted to 10 cm diameter plastic pots, filled with acid-washed sand. In each pot ten seedlings were established. Later on, thinning was performed and 5 healthy plants were maintained in each pot. Plants grown in pots were kept under natural illuminated conditions (General climatic conditions such as humidity, day/night temperature were recorded during the experiment). To fulfill nutrient requirement of growing plants, all the pots were supplied with 50 mL of Raukura’s nutrient solution [9] every day. The salts used to make up the nutrient solution were as follows: Macronutrient stock solution A (g∙L−1): Mg (NO3)2 6H2O, 4.94; NH4NO3, 8.48; KNO3, 2.28. Macronutrient stock solution B (g∙L−1): KH2PO4, 2.67; K2HPO4, 1.64; K2SO4, 6.62; Na2SO4, 0.60; NaCl, 0.33. Micronutrient supplement (mg∙L−1): H3BO3, 128.80; CuCl2∙2H2O, 4.84; MnCl2∙4H2O, 81.10; (NH4)6∙Mo7O24∙4H2O, 0.83; ZnCl2, 23.45; C6H5FeO7∙5H2O, 809.84. The applied dilute solution was prepared by mixing 100 mL of each of the macronutrient stock solution with 50 mL of the micronutrient supplement and diluting to 4.5 L with DDW. The pH was maintained at 6.0 by adding H2SO4 or KOH. 200 mM NaCl was also applied along with nutrient solution. To avoid osmotic shock, NaCl concentration was increased by 25 mM every two days until the desired concentration of 200 mM was achieved. Each pot was considered as one replicate and the treatment for each genotype was repeated three times.
Response of plants to salt stress was assessed at 60 DAS in terms of growth parameters viz. plant height, fresh weight, dry weight, number of leaves, leaf area per plant (LA); and physiological and biochemical parameters viz. carbonic anhydrase activity, nitrate reductase activity, leaf chlorophyll content (Chl), leaf relative water content (LRWC), lipid peroxidation, and activities of antioxidant enzymes [superoxide dismutase (SOD), peroxidase (POX), and catalase (CAT)].
2.2. Measurement of Growth Attributes
Growth of plants in response to NaCl was assessed in terms of plant height, fresh weight, dry weight, number of leaves, and leaf area per plant (LA). Dry weight was recorded by drying the plants at 80˚C for 24 h.
The leaf area of about 10% of leaves (LA1) was measured by tracing the leaves on a graph paper and dry weight of these leaves was recorded (W1). Leaf dry weight of total leaves per plant (W2) was registered. Leaf area per plant was calculated using the following formula [10] .
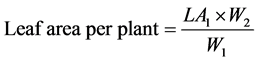
2.3. Determination of Physiological and Biochemical Parameters
The activity of CA (E.C. 4.2.1.1) was measured using the method as described by Dwivedi and Randhawa [11] . The enzyme was expressed as µM CO2 kg−1 leaf FW s−1. Activity of NR (E.C. 1.6.6.1) was estimated by the intact tissue method developed by Jaworski [12] . Activity of NR was expressed as µM 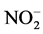 g−1 leaf FW h−1. Total chlorophyll content in the leaves was determined using the method of Lichtenthaler and Buschmann [13] . The photosynthetic pigment, thus measured, was expressed as mg∙g−1 leaf FW. Leaf relative water content (LRWC) was measured by adopting the method of Yamasaki and Dillenburg [14] . Fresh weight (FW), dry weight (DW) and turgid weight (TW) of leaves was measured and LRWC was calculated using the equation below.
g−1 leaf FW h−1. Total chlorophyll content in the leaves was determined using the method of Lichtenthaler and Buschmann [13] . The photosynthetic pigment, thus measured, was expressed as mg∙g−1 leaf FW. Leaf relative water content (LRWC) was measured by adopting the method of Yamasaki and Dillenburg [14] . Fresh weight (FW), dry weight (DW) and turgid weight (TW) of leaves was measured and LRWC was calculated using the equation below.
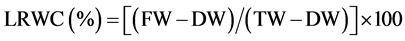
Lipid peroxidation was estimated by the content of thiobarbituric acid reactive substances (TBARS) as described by Cakmak and Horst [15] . TBARS content are expressed as nM g−1 leaf FW.
2.4. Assay of Antioxidative Enzymes
Leaf tissues were homogenized with three volumes (w/v) of an ice-cold extraction buffer (50 mM Tris-HCl, pH 7.8, 1 mM EDTA, 1 mM MgCl2 and 1.5% (w/w) polyvinyl pyrrolidone). The homogenate was centrifuged at 15,000 g for 20 min at 4˚C. The supernatant was used as the crude extract for the assay of enzyme activities.
Activity of SOD (E.C. 1.15.1.1), POX (E.C. 1.11.1.7) and CAT (E.C. 1.11.1.6) was determined according to Beauchamp and Fridovich [16] , Upadhyaya et al. [17] and Cakmak and Marschner [18] , respectively.
2.5. Statistical Analysis
Each pot was considered as one replicate and the treatment for each genotype was repeated three times. The data were analyzed statistically with SPSS-17 statistical software (SPSS Inc., Chicago, IL, USA). When F value was found to be significant at 5% level of probability, least significant difference (LSD) was calculated. Values were expressed as means ± standard deviation (SD).
3. Results and Discussion
3.1. Effect of Salt Stress on Growth Attributes of Ten Genotypes of Tomato
In the present experiment response of ten genotypes to 200 mM NaCl was tested to evaluate salt tolerant and salt sensitive genotypes. Perusal of the data show that the tested genotypes responded differently to NaCl stress (Table 1 and Table 2). Regarding plant height, genotype “Plitz” gave highest value for plant height whereas, genotype “Grace” gave the lowest value for plant height among the tested genotypes (Table 1). When the effect of salt stress on leaf number and leaf area was tested, BL-1076 had maximum number of leaves and leaf area. However, BL-1076, in term of leaf number, showed parity with Grace, Imperial, PakmoreVF and Bonus F1 genotypes. Lowest number of leaves was recorded in the Queen (Table 1). However, shoot fresh and dry weight were the highest in BL-1076, whereas the lowest values for shoot fresh and dry weight were registered in Queen genotype. The same pattern was also recorded for root fresh and dry weight (Table 2). Thus, on the basis of growth parameters BL-1076 genotype was found salt tolerant while Queen was registered as salt sensitive genotype. The present results are in agreement with the findings of El Sayed et al. [19] , who reported that presence
Table 1. Effect of 200 mM NaCl on plant height, number of leaves and leaf area of ten genotypes of tomato (Lycopersicon esculentum Mill.).
Average of three determinations is presented with LSD at 5%; ± indicating S.D.
Table 2. Effect of 200 mM NaCl on shoot and root dry weight and fresh weight of ten genotypes of tomato (Lycopersicon esculentum Mill.).
Average of three determinations is presented with LSD at 5%; ± indicating S.D.
of NaCl in the growth medium significantly decreases growth attributes of tomato. As shown by the results, 200 mM NaCl caused highest reduction in the activities of CA and NR enzymes (Figure 1(a) and Figure 1(b)) in the genotype Queen, which enabled the plants with reduced photosynthetic capacity and nitrogen assimilation which ultimately culminated in reduced dry matter accumulation (Table 2). However, reverse is true for the genotype BL-1076 which accumulated more dry matter compared to Queen. It has been observed that higher levels of salinity cause generation of ROS. In the present investigation, presence of NaCl enhanced the level of TBARS [20] and lowered the activities of antioxidant enzymes in genotype Queen, whereas, lower TBARS and higher activities of antioxidant enzymes were noticed in genotype BL-1076. Thus, it is clear that genotype Queen was sensitive while BL-1076 was tolerant to the NaCl stress. Response of tomato genotypes to salinity
Figure 1. Effect of 200 mM NaCl on the enzyme activities of carbonic anhydrase (a) and nitrate reductase (b), chlorophyll content (c) and leaf relative water content (d) of ten genotypes of tomato (Lycopersicon esculentum Mill.). CA: carbonic anhydrase; NR: nitrate reductase; LRWC: leaf relative water content. Average of three determinations is presented with LSD at 5%, and T bars indicating S.D.
revealed that the genotypes had a wide range of variability for all plant growth traits (Alsadon et al., 2013).
3.2. Effect of Salt Stress on Physiological and Biochemical Attributes
Salt tolerance capacity of the selected genotypes was also tested in terms of physiological and biochemical parameters. The results exhibit that genotype BL-1076 gave highest values for the activities of CA and NR enzymes under 200 mM NaCl stress. However, Queen gave lowest values for these two enzymes (Figure 1(a) and Figure 1(b)). Chlorophyll content and relative water content are the two important parameters in determining the salt tolerance capacity of plants. The genotype BL-1076 again proved best and gave highest leaf chlorophyll and water content under salt stress, while Queen performed worst (Figure 1(c) and Figure 1(d)). Nezhad et al. [21] observed that salt stress decreased chlorophyll content in the leaves of tomato plant. On the other hand, Sayed et al. [19] observed an increase in chlorophyll content under salt stress. A decrease in CA and NR activities and leaf chlorophyll and relative water content was recorded in NaCl-stressed leaves of mustard [22] -[24] . Salt stressed plants accumulated higher Na+ and resulted in reduction of the water status of leaves, it may be responsible for the higher levels of TBARS (Figure 2(a)) that slowed down the activities of CA and NR which might have led to reduced leaf chlorophyll content (Figure 1(c)). Reduction of Chl content was probably due to the toxic effect of salinity on the instability of protein complexes and increase in the activity of chlorophyll-de- grading enzyme chlorophyllase [25] leading to destruction of chlorophyll. Effect of salt stress on lipid peroxidation was tested in term of content of TBARS. Highest membrane damage was registered in Queen while least in BL-1076 which gave highest and lowest TBARS content respectively (Figure 2(a)). In order to counter stress induced oxidative stress plant are equipped with a system of antioxidative enzymes such as SOD, POX and CAT. In the present study, we noticed that among the ten genotypes tested, BL-1076 showed the highest activity of SOD, POX and CAT, however, Queen gave the lowest values for these three antioxidative enzymes (Figure 2(b) and
Figure 2. Effect of 200 mM NaCl on TBARS content (a) activities of SOD and POX (b) and CAT activity (c) of ten genotypes of tomato (Lycopersicon esculentum Mill.). TBARS: Thiobarbituric acid reactive substances; SOD: superoxide dismutase; POX: peroxidase; CAT: catalase. Average of three determinations is presented with LSD at 5%, and T bars indicating S.D.
Figure 2(c)). Therefore, higher level of TBARS in Queen can be explained on the basis of lower activities of antioxidant enzymes which were unable to scavenge ROS and thus higher levels of TBARS were reported in the Queen. However, reverse is true for the genotype BL-1076 which gave higher activities of antioxidant enzymes leading to lower TBARS content. Therefore, BL-1076 genotype was again found salt tolerant while Queen as salt sensitive. The differences in genotypes in respect of physiological and biochemical parameters may be ascribed to the variation in the genetic makeup of the genotypes.
4. Conclusion
Present investigation was carried out to evaluate salt tolerant and salt sensitive genotypes of tomato through assessing growth and physio-biochemical attributes. The results show that all the ten tested genotypes exhibited a wide range of variability in growth, physiological and biochemical attributes in response to salinity. Genotype BL-1076 exhibited higher values of antioxidant enzymes that assisted plants in maintaining optimum activities of CA and NR enzymes, leaf chlorophyll content and leaf relative water content. All these together contributed to improved growth of the plants of BL-1076 genotype. However, reverse is true for the genotype Queen which exhibited lower activities of antioxidant enzymes coupled with lower values of growth, physiological and biochemical characteristics. Thus, the results confirm that BL-1076 was salt tolerant while Queen salt sensitive. Variation in the performance of tested genotypes under salinity leads to the conclusion that these parameters were governed by the genetic makeup of the genotypes. To put all in a nut shell the genotypes can be arranged in the following order of their decreasing salt tolerance capacity: BL-1076 > Trust > Imperial > Tanshet star > PakmoreVF > L 26 > Plitz > Bonus F1 > Grace > Queen. As in the present investigation a genetic variability within a species was recorded which was considered as a valuable tool for plant breeding studies. Therefore, the evaluated genotypes can be used for plant breeding projects of tomato to improve fruit yield, quality and resistance to salt stress.
Acknowledgements
The financial support (Project no. S-0105-1436) by Deanship of Scientific Research (DSR), University of Tabuk, Saudi Arabia is gratefully acknowledged. The author would also like to thank the Head of the Department of Biology, Faculty of Sciences, University of Tabuk for providing necessary facilities and thanks are also to Saudi Digital Library and University Library for providing relevant and updated literature.
Cite this paper
M. NasirKhan, (2016) Growth and Physiological Attributes of Tomato (Lycopersicon esculentum Mill.) Genotypes as Affected by NaCl Stress. American Journal of Plant Sciences,07,453-460. doi: 10.4236/ajps.2016.73039
References
- 1. Ilahy, R., Hdider, C., Lenucci, M.S., Tlili, I. and Dalessandr, G. (2011) Phytochemical Composition and Antioxidant Activity of High-Lycopene Tomato (Solanum lycopersicum L.) Cultivars Grown in Southern Italy. Scientia Horticulturae, 127, 255-261.
http://dx.doi.org/10.1016/j.scienta.2010.10.001 - 2. Pinela, J., Barros, L., Carvalho, A.M. and Ferreira, I.C. (2012) Nutritional Composition and Antioxidant Activity of Four Tomato (Lycopersicon esculentum L.) Farmer’s Varieties in Northeastern Portugal Homegardens. Food and Chemical Toxicology, 50, 829-834.
http://dx.doi.org/10.1016/j.fct.2011.11.045 - 3. Taveira, M., Ferreres, F., Gil-Izquierdo, A., Oliveira, V., Valentäo, P. and Andrade, P.B. (2012) Fast Determination of Bioactive Compounds from Lycopersicon esculentum Mill. Leaves. Food Chemistry, 135, 748-755.
http://dx.doi.org/10.1016/j.foodchem.2012.05.016 - 4. Meckna, B., Reimers, K. and Mark, A. (2012) Antioxidant Capacity of Tomato Paste Is Stable during Growing Season and Shelf-Life. The FASEB Journal, 26, 1017.6.
- 5. Glenn, E.P., Brown, J.J. and Khan, M.J. (1997) Mechanisms of Salt Tolerance in Higher Plants. In: Basra, A.S. and Basra, R.K., Eds., Mechanisms of Environmental Stress Resistance in Plants, Harwood Academic Publishers, The Netherlands, 83-110.
- 6. Munns, R., James, R.A. and Läuchli, A. (2006) Approaches to Increasing the Salt Tolerance of Wheat and Other Cereals. Journal of Experimental Botany, 57, 1025-1043.
http://dx.doi.org/10.1093/jxb/erj100 - 7. Zhu, J.K. (2001) Plant Salt Tolerance. Trends in Plant Science, 6, 66-71.
http://dx.doi.org/10.1016/S1360-1385(00)01838-0 - 8. Foyer, C.H. and Noctor, G. (2000) Oxygen Processing in Photosynthesis: Regulation and Signaling. New Phytologist, 146, 359-388.
http://dx.doi.org/10.1046/j.1469-8137.2000.00667.x - 9. Smith, G.S., Johnston, C.M. and Cornforth, I.S. (1983) Comparison of Nutrient Solutions for Growth of Plants in Sand Culture. New Phytologist, 94, 537-548.
http://dx.doi.org/10.1111/j.1469-8137.1983.tb04863.x - 10. Watson, D.J. (1958) The Dependence of Net Assimilation Rate on Leaf Area Index. Annals of Botany, 22, 37-54.
- 11. Dwivedi, R.S. and Randhawa, N.S. (1974) Evaluation of Rapid Test for Hidden Hunger of Zinc in Plants. Plant and Soil, 40, 445-451.
http://dx.doi.org/10.1007/BF00011531 - 12. Jaworski, E.J. (1971) Nitrate Reductase Assay in Intact Plant Tissues. Biochemical and Biophysical Research Communications, 43, 1274-1279.
http://dx.doi.org/10.1016/S0006-291X(71)80010-4 - 13. Lichtenthaler, H.K. and Buschmann, C. (2001) Chlorophylls and Carotenoids: Measurement and Characterization by UV-VIS Spectroscopy. In: Wrolstad, R.E., Acree, T.E., An, H., Decker, E.A., Penner, M.H., Reid, D.S., Schwartz, S.J., Shoemaker, C.F. and Sporns, P., Eds., Current Protocols in Food Analytical Chemistry, John Wiley and Sons, New York, F4.3.1-F4.3.8.
- 14. Yamasaki, S. and Dillenburg, L.C. (1999) Measurements of Leaf Relative Water Content in Araucaria angustifolia. Revista Brasileira De Fisiologia Vegetal, 11, 69-75.
- 15. Cakmak, I. and Horst, J.H. (1991) Effects of Aluminum on Lipid Peroxidation, Superoxide Dismutase, Catalase, and Peroxidase Activities in Root Tips of Soybean (Glycine max). Physiologia Plantarum, 83, 463-468.
http://dx.doi.org/10.1111/j.1399-3054.1991.tb00121.x - 16. Beauchamp, C. and Fridovich, I. (1971) Superoxide Dismutase: Improved Assays and an Assay Applicable to Acrylamide Gels. Annals of Biochemistry, 44, 276-287.
http://dx.doi.org/10.1016/0003-2697(71)90370-8 - 17. Upadhyaya, A., Sankhla, D., Davis, T.D., Sankhla, N. and Smith, B.N. (1985) Effect of Paclobutrazol on the Activities of Some Enzymes of Activated Oxygen Metabolism and Lipid Peroxidation in Senescing Soybean Leaves. Journal of Plant Physiology, 121, 453-461.
http://dx.doi.org/10.1016/S0176-1617(85)80081-X - 18. Cakmak, I. and Marschner, H. (1992) Magnesium Deficiency and High Light Intensity Enhance Activities of Superoxide Dismutase, Ascorbate Peroxidase, and Glutathione Reductase in Bean Leaves. Plant Physiology, 98, 1222-1227.
http://dx.doi.org/10.1104/pp.98.4.1222 - 19. El-Sayed, H.E.S.A., Baziad, S.A.M. and Basaba, R.A.A.S. (2015) Application of Exogenous Ascorbic Acid on Tomato (Solanum lycopersicum L.) Seeds under NaCl Salinity Stress. International Journal of Current Research in Biosciences and Plant Biology, 2, 33-46.
- 20. Hassan, M.A., Fuertes, M.M., Sánchez, F.J.R., Vicente, O. and Boscaiu, M. (2015) Effects of Salt and Water Stress on Plant Growth and on Accumulation of Osmolytes and Antioxidant Compounds in Cherry Tomato. Notulae Botanicae Horti Agrobotanici, 43, 1-11.
http://dx.doi.org/10.15835/nbha.43.1.9793 - 21. Nezhad, S.F., Ehsanpour, A.A. and Hosseini, S.M. (2011) Effect of Salt Stress on Acid Phosphatase Activity and Phosphorus Content of Lycopersicon peruvianum L. under in Vitro Culture. Malaysian Applied Biology, 40, 7-11.
- 22. Khan, M.N., Siddiqui, M.H., Mohammad, F. and Naeem, M. (2012) Interactive Role of Nitric Oxide and Calcium Chloride in Enhancing Tolerance to Salt Stress. Nitric Oxide, 27, 210-218.
http://dx.doi.org/10.1016/j.niox.2012.07.005 - 23. Reddy, M.P. and Vora, A.B. (1986) Changes in Pigment Composition, Hill Reaction Activity and Saccharides Metabolism in Bajra (Pennisetum typhoides S&H) Leaves under NaCl Salinity. Photosynthetica, 20, 50-55.




