E-Health Telecommunication Systems and Networks
Vol.03 No.03(2014), Article ID:49724,9 pages
10.4236/etsn.2014.33005
eHealth Management Platform for Screening and Prediction of Down’s Syndrome in the Republic of Panama
Juan Saldaña, Miguel Vargas-Lombardo
Software Engineering Department, Technological University of Panama, Panama City, Panama
Email: juan.saldana@utp.ac.pa, Miguel.vargas@utp.ac.pa
Copyright © 2014 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 27 July 2014; revised 25 August 2014; accepted 9 September 2014
ABSTRACT
Through engineering projects, we have integrated software engineering, geographical information systems and HL7 standard to propose a model of an eHealth management platform for Down’s syndrome screening, replicable in all the country. It will use real time sample information acquired from the local population and will geographically reference this information in the territory of Panama for future research.
Keywords:
Project Engineering, Software Engineering, GIS, eHealth, Management Platform, HL7, Trisomy 21, Sample, Screening, Detection

1. Introduction
In many countries like Panama, a lot of medical information is not saved electronically. It brings deficiency to the health system of the country affecting different areas of health care. It makes necessary the use of eHealth management platforms. For this reason, we want to focus our work in one of the illness that is actually affected by the lack of information which is the Down’s syndrome also called trisomy 21.
In the health community, the hospital information systems are not developed with standards that help to manage the patient’s clinical information [1] -[3] . The clinical information is scattered around the country in different administrative entities and private and public hospitals.
In order to get clinical data about trisomy 21, we need to access, collect and analyze data from various health entities. Because of that, a lot of patients are wrongly diagnosed or receive their treatments later than due.
In this document we make a proposal for the analysis, model and development of an eHealth management platform for the antenatal screening for Down’s syndrome in the republic of Panama.
The rest of the document is structured as follow: Section two is a state of the art about the trisomy 21 and the current trends in antenatal screening for trisomy 21. Section three explains the technologies that are going to be used to develop the eHealth management platform. Section four is a revision of others eHealth platforms. Section five explains the proposed eHealth management platform’s architecture. The next section presents the conclusions of our work.
2. Trisomy 21
2.1. Aneuploidy Trisomy 21
Trisomy 21 is an Aneuploidy where the fetus shows a genetic alteration having 3 chromosomes 21 [4] [5] . This trisomy is one of the major causes of deficiencies or physical disabilities in children, premature deaths that take place before birth, situation that many mothers are unaware. This chromosomal disorder causes various physical deformities, hearth defects, organ malformations, mental retardation, thyroid disorder and diseases like Alzheimer’s. Trisomy 21 responsible for Down’s syndrome is the most frequent aneuploidy
2.2. Probabilistic Method for Trisomy 21 Detection
The Screening is a probabilistic technique applied to a population to calculate the risk or probability that the fetus suffer a particular disease.
In the screening, first serum and biochemical markers [3] are established and compared with historical medians references values of the population. When the test results of the patient and the MOM or Multiple of Median of the markers [4] have different values, the test is considered positive.
The effectiveness of this method is measured in terms of its detection rate (DR) which it is better the greater its value, and depending on the detection rate of false positives (FPR) [6] [7] . These two variables are affected by what is known as the cutoff, making the effectiveness of the test depends off this value.
These rates should be 75% DR and 3% FPR based on the standard set in 2008 by the National Screening Committee with aspirations of reaching 90% and 2% respectively [4] and even reduced to 1% and 1.5%.
The screening is performed in the second and first trimester being the first trimester the most difficult to realize because of various reason being one of them the lack of sampling data to perform the test.
Initially, second trimester reached a detection rate of 80%. Then in 2000, it began to be performed first trimester screening with 85% detection rate and false positive of 5%.
For the first trimester trisomy 21 screening, it is necessary to perform a more effective detection taking in consideration some mother’s characteristics like her weight, her ethnicity, if she has diabetes or if is smoke. These factors could affect the result of the test so it needs to be corrected.
3. eHealth Management Platform Technologies
3.1. Component Based Development in the Cloud
Component based development in the cloud is also known as Software as a Service. It provides to us a lot of benefits like access to resources as they are needed and out of the box, development time and cost reduction [8] - [11] . These are important values for the proposed platform. There are important component like the geographical system that will be incorporated in our design. This component is widely used, tested, secure and free.
3.2. HL7
A common disadvantage that most of the hospital information systems have for the clinical information management is lack of interoperability between them. Health Level Seven HL7 [1] [3] [10] [12] -[15] and Open HER [12] [16] [17] are international standards to manage clinical information electronically promoting the interoperability.
3.3. GIS
Geographical information systems are systems that use latitude and longitude allowing data geo reference [1] [3] [18] . This geographical reference applied to health information data allows the development of useful tools. For example, we could detect geographic causes of diseases or follow the spread of infectious diseases with updated online information [18] .
3.4. Software Engineering and eHealth
The term eHealth is fairly new. As mentioned in [13] [19] [20] , it can be defined as a new paradigm for the management of medical information that involves technology aspects to access, share and store electronic patient’s information, administrative and operational processes in health institutions and the interoperability between hospital information systems. But it is more than that, It also means a culture change, a way of thinking, seeing and analyzing the entire domain in order to improve patient’s health care.
4. Comparison of Current eHealth Platforms with PLAGETRI21
The use of web platforms in the e-health field is becoming more common. These are used in different areas in the health domain to achieve different objectives. For example, Peixoto [21] is an eHealth platform, used by technical staff, administrative staff and clinical team. It manages preventive actions to keep running the infrastructure of a hospital information system based on action estimation models.
Another platform related with aneuploidy for the risk calculation of trisomy 21 is SBP Software [22] . Unlike PLAGETRI21, SBP Software does not calculate the MoM. To perform the value calculation of each indicator, the MoM must be calculated using some other tool or its used generics MoMs that are calculated in other countries. Therefore, the results will be based on non-local data. The platform also has drawbacks because of the small amount of information variables stored for each patient, decreasing the possibility of future studies and research.
There also are software packages like Typolog Software and SsdwLab [4] [23] . The system architecture of Typolog software stores the information locally, restricting the possibility of using the data for sampling in future analysis of variables correlation.
SBP Software does not include important variables for the risk calculation method like the mother ethnic, weight and others. The sampling data used it is not collected from the Panamanian population. SsdwLab is not multiplatform; it has not interoperability features and it has the same disadvantage of SBP Software.
In Table 1, it can be appreciated a brief comparison of these platforms with PLAGETRI21.
The platform PLAGETRI21 is designed to meet the information needs of our country, to improve the current method of trisomy 21 risk calculation, implementing the first trimester test and provide information for future research about these diseases in Panama.
5. Architecture Components of the eHealth Management Platform for Trisomy 21 Risk Prediction in the Republic of Panama
For the platform design, we have adopted the goal-directed design method created by Allan Cooper [21] [24] . The design’s idea is to be as simple as possible focusing on the main goal and what the medical staff needs to achieve it. Focusing on which are the functional features that will solve the current deficiencies of the methods for estimating a woman’s risk of having a pregnancy associated with Down’s syndrome in our country.
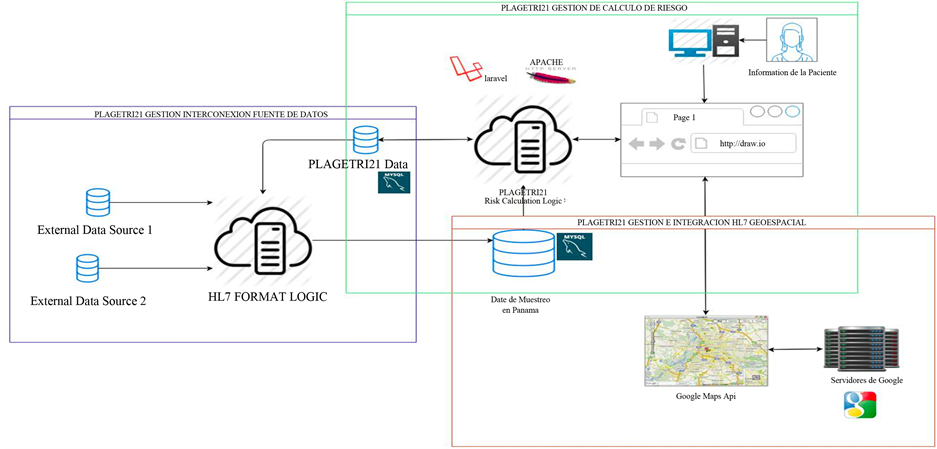
The proposed architecture can be appreciated in Figure 1. It is composed of three main components: thse risk
Table 1. Brief platform comparison.
Figure 1. PLAGETRI21 eHealth management platform architecture.
calculation management component, database interconnection component and the geospatial-referencing component.
5.1. Trisomy 21 Risk Calculation Management Component
The eHealth Management Platform PLAGETRI21 is based on the calculation risk method of Likelihood, published by Palomaki and Haddow [25] combining the a priori risk for maternal age obtained from the meta-anal- ysis Cuckle HS, Wald NJ and Thompson [26] , with the likelihood obtained from combining the MoM of the different markers used in each profile. The MoM is calculated using the multivariate normal distribution published by Reynolds and Penney MD TM [27] .
This calculation method is applied by important programs currently used by healthcare companies such as Roche to estimate the patient’s risk of trisomy 21 [5] [8] [22] [28] .
To explain how PLAGETRI21 works, it is important to first understand some concepts. PLAGETRI21 implement a not invasive diagnostic method. The prenatal diagnosis of the chromosomal defects for trisomy 21 detection usually implements invasive procedures like the amniocentesis and chorionic villus samples [3] . This type of test presents some danger to the fetus because of their nature.
The platform determines which patient has a major risk of having this pathology from the one is sane.
Once the risk is calculated, the patient with high risk should receive a diagnostic method to detect if the patient really has the disease.
With this platform, we look for first improve the results from the currents diagnostics method used in Panama, working with a real sampling with data from patients in the country and second improve the procedures to realize first trimester screening test.
The screening procedure compares the patient result with the MoMs of the sampling. When the comparison is completed, the analysis result could be located in 4 groups: true positives, true negatives, the false positives and the false negatives [20] [29] . The idea is to detect a high rate of true positives cases with the small rate of false negatives.
The trisomy 21 risk is calculated dividing 1 by the inverse of the probability. (Equation (1): Trisomy 21 Risk Calculation Formula).
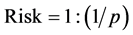 (1)
(1)
To make the calculation, a mathematical and statistical process is done with following steps:
· Risk estimation based in the maternal age.
· Markers standardization.
· Multiple of medians (MoM) calculation.
· Weight and correction factors adjustments.
Maternal weight corrections.
Covariates corrections.
· Likelihood ratio estimation.
· Risk estimation.
5.1.1. Maternal Age Risk Estimation
This is the first marker that will be used in the platform with the method proposed by Cuckle et al. [6] [22] [30] . Currents software vendors use this risk estimation method.
The equation for the risk calculation can be appreciated below. (Equation (2): Maternal Age Risk Estimation).
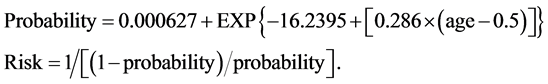 (2)
(2)
For the age evaluation, there is also a risk known as fetus prior affection [20] that establishes that the risk that a patient has of presenting trisomy 21 increase and it is higher than the risk calculate based on the age, when this patient has already presented a trisomy 21 case before.
The intrauterine lethality [20] is also one important variable that needs to be taken in consideration based on the age of the mother. This variable is calculated based on the aneuploidy observed versus the expected, from mothers with different age. This correction factor should be used when you want to present the trisomy 21 risk that the fetus has in a given trimester. In this case according to [6] for trisomy 21 the correction rate is as follows Table 2.
5.1.2. Standardization of Markers Values
The markers values are variables that follow a Gaussian distribution. These values will vary depending on the gestational age and specific aspects of the mother such as age, race and weight [31] .
It is necessary to establish the median of the markers for each gestation week in order to make the transformation of the markers and thereby reduce the effect of variation that occurs in each week of pregnancy. After these medians are known, it can be set multiples of medians MoM for each marker.
The eHealth management platform must perform the calculation based on medians that will be determined by analyzing the values of women who have or not positive cases of trisomy. These values will be provided by the data sources of health institutions that are part of the project. The fact of having local data sources let us include in the study specific characteristics of the population or region that could influence the median calculations. These characteristics cannot be analyzed with data from foreign woman and other regions.
5.1.3. Multiple of Median Calculation (MoM)
The MoMs are calculated by the division of the value of the marker obtained with the test, between the medians calculated for this marker. (Equation (3): Multiple of Median Calculation).
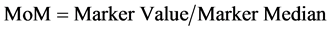 (3)
(3)
5.1.4. Adjustments Based on the Weight Correction Factors
As mentioned above, specific properties of the mother like the weight for example, affects the markers values so it is necessary to correct them. The weight correction can be done applying two methods: The exponential method: (Equation (4): Exponential Maternal Weight Correction).
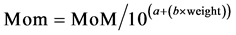 (4)
(4)
And the lineal reciprocal method: (Equation (5): Lineal Maternal Weight Correction).
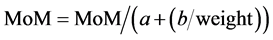 (5)
(5)
These two correction formulas use coefficients that are shown in the Table 3:
5.1.5. Correction Factor or Covariate FC
In addition to the maternal weight corrections, it is also necessary to make corrections of the markers values
Table 2. Correction rate of trisomy 21 based on the trimester.
Table 3. Coefficients for the weight regression formulas [16] .
based on factors such as race, if the patient has diabetes or smokes and the fetal sex [17] .
The median for the correction factor for each genetic group are shown in Table 4 and the formula for these corrections is the following: (Equation (6): Factor Corrections of the MoM).
 (6)
(6)
5.1.6. Likelihood Ratio Calculation for Trisomy 21
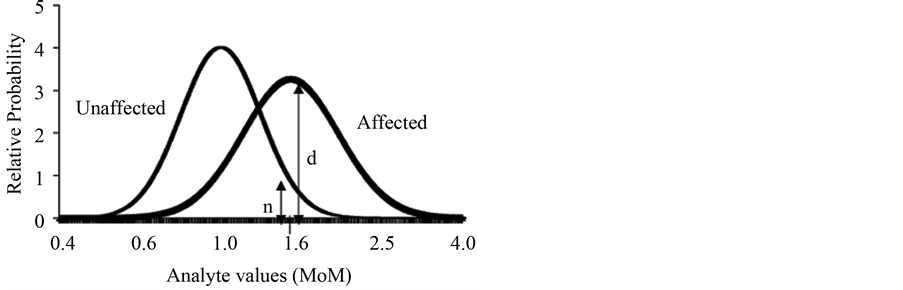
Likelihood calculation is performed based on the multivariate normal distribution [5] [32] using the population parameters. The population parameters consist in three values: the median, the standard deviation of each marker and the variation coefficient between pairs of markers (if it is used more than one marker) for both the healthy and affected population. The likelihood ratio is calculate dividing (d + n) /n as shown in Figure 2.
5.1.7. Pregnancy without Aneuploidy
The standard deviation of the markers varies from the trimester and correction factors used. For the population that doesn’t present aneuploidy, the standard deviation will be found within the following limits show in Table 5.
5.1.8. Gestations with Aneuploidy
The population’s sampling values of cases with aneuploidy is much lower than the unaffected. Because of the small amount of data from affected cases, most of the current test uses generic data from research made in other countries. The eHealth management platform PLAGETRI21 will record information from many local [33] institutions, allowing us to make the test using data values from the Panamanian population.
For affected cases the variable values must be within the following limits show in Table 6.
The correlation coefficients for gestations without aneuploidy should be within show in Table 7.
5.1.9. Trisomy 21 Risk Estimation
Trisomy 21 risk is calculated multiplying the maternal age estimated risk by the calculated likelihood of the trisomy [25] [27] [33] . (Equation (7): Trisomy 21 Risk).
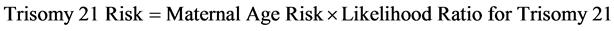 (7)
(7)
5.2. eHealth Management Platform for the Interconnection with Data Sources and HL7 Interoperability.
The eHealth management platform PLAGETRI21 architecture is based on the architecture presented in [9] [34] which allow the interoperability between many hospital information systems.
PLAGETRI21 will be able to interact using the HL7 standard with many data source of clinical information. It will collect sampling data necessary to calculate more efficiently the trisomy 21 risk and make future research in this domain.
Figure 2. Likelihood ratios.
Table 4. Median for AFP, hCG and uE3 for each genetic group [16] .
Table 5. Deviation range in a healthy population [32] .
Table 6. Median and standard deviation values for affected cases [32] .
Table 7. Correlation coefficients for not affected gestations [20] .
The interconnection with other data sources involves security and privacy of the information like is presented by Geissbuhler in [13] . The data that is extracted from the different data sources will include general information of the patient like age, sex, ethnic, blood type, place of birth, residence. Specific fields like name, last names, and personal ID will not be included in the sampling data source. The information will be copied from the data source without modify the original information. It will be exported and saved to the sampling database using a web service.
All the process consists in a group of transaction that will implement HL7 like it is implemented in [4] [35] to guarantee an efficient and reliable information management.
5.3. eHealth Management Platform and Spatial HL7 Integration with the Geographical Information System
The eHealth management platform PLAGETRI21 will implement the geospatial interoperability standard proposed in [1] [3] . The inclusion of geographical information to the platform will enable the clinical information georeferencing by birth place, residence place and location where the patient receives the medical care, allowing us to save and visualize in a geographical system, information required to in the future research deeper about this affection. A geographical information system component existing in the cloud will be integrated with the platform to have access to the geographical data and the component features, benefits and technology.
6. Conclusions
· The development of PLAGETRI21 will collect extremely useful data to make more reliable tests of trisomy 21 and it will allow performing the first trimester screening test for trisomy 21 in Panama.
· The database that PLAGETRI21 manage is a really important element to the future research in the field of Down’s syndrome in Panama.
· This eHealth Management Platform will be a prototype for future works in medical areas like epidemiology for example, for the control of infectious diseases in the country as presented in [36] .
· Implementing standards like HL7, we open the door for the future integration of PLAGETRI21 with other platforms that will probably extend the initial functionality and objectives of the platform.
References
- Esri, A. (2011) HL7 and Spatial Interoperability Standards for Public Health and Health Care Delivery.
- Fernández, A. and Oviedo, E. (2011) E-Health in Latin America and the Caribbean.
- Fonseca, J.M., Mora, A.D. and Barroso, P. (2007) The Web and the New Generation of Medical Information Systems. Journal Outcome Prediction in Cancer, 391-414.
- Typolog, S. (2014) Typolog Software. Online. http://www.typolog.de/SharedDatabase.html
- Benn, P.A. (2002) Advances in Prenatal Screening for Down Syndrome: I. General Principles and Second Trimester testing. Clinica Chimica Acta, 323, 1-16
- Fuentes, M.N. (2011) Screening de aneuploidías del primer trimester. 1-18.
- Gyselaers, W.J.A., Vereecken, A.J., Van Herck, E.J.H., Straetmans, D.P.L., Martens, G.E.I., de Jonge, E.T.M., Ombelet, W.U.A.M. and Nijhuis, J.G. (2004) Screening for Trisomy 21 in Flanders: A 10 Years Review of 40.490 Pregnancies Screened by Maternal Serum. European Journal of Obstetrics & Gynecology and Reproductive Biology, 115, 185- 189. http://dx.doi.org/10.1016/j.ejogrb.2003.12.014
- Kitchenham, B., Brereton, O.P., Budgen, D., Turner, M., Bailey, J. and Linkman, S. (2009) Systematic Literature Reviews in Software Engineering—A Systematic Literature Review. Information and Software Technology, 51, 7-15. http://dx.doi.org/10.1016/j.infsof.2008.09.009
- Khalifa, M. (2013) Barriers to Health Information Systems and Electronic Medical Records Implementation. A Field Study of Saudi Arabian Hospitals. ProcediaComput. Sci., 21, 335-342.
- Greenfield, J. (2013) Designing and Implementing an HL7 Software Factory. 1-7.
- Zrnec, A. (2011) Software Engineering in the Cloud for Reducing the Application Time-to-Market. Vol. 78, 123-127.
- Dande, A.C. and Sunder, G. (2009) Suitability of HL7 v3 in Building an EHR System. 3-6.
- Geissbuhler, A. (2013) Lessons Learned Implementing a Regional Health Information Exchange in Geneva as a Pilot for the Swiss National eHealth Strategy. International Journal of Medical Informatics, 82, e118-e124. http://dx.doi.org/10.1016/j.ijmedinf.2012.11.002
- Aguilar, R. and López, D. (2009) Guia de Implementación HL7 parasistemas de notificaciónobligatoria en Salud Púb- lica en Colombia. 13-32.
- Tudor, C.R.P.H. (2011) Project eSanté Architecture and Security of a National eHealth Platform.
- Vargas, J.D.L., Velez, D.P. and Bustamante, F.P. (2013) Estudio y desarrollo de un sistema que implementa el estándar hl7 de registros médicos generales y envia comentarios por mensajes de texto.
- Duftschmid, G., Rinner, C., Kohler, M., Huebner-Bloder, G., Saboor, S. and Ammenwerth, E. (2013) The EHR- ARCHE Project: Satisfying Clinical Information Needs in a Shared Electronic Health Record System Based on IHE XDS and Archetypes. International Journal of Medical Informatics, 82, 1195-1207. http://dx.doi.org/10.1016/j.ijmedinf.2013.08.002
- Granell, C., Fernández, Ó.B. and Díaz, L. (2014) Geospatial Information Infrastructures to Address Spatial Needs in Health: Collaboration, Challenges and Opportunities. Future Generation Computer Systems, 31, 213-222. http://dx.doi.org/10.1016/j.future.2013.04.002
- Cooper, A., Reimann, R. and Cronin, D. (2004) About Face 3.0: The Essentials of Interaction Design, 3, 223-225.
- Mettler, T. and Eurich, M. (2012) A “Design-Pattern”-Based Approach for Analyzing e-Health Business Models. Future Generation Computer Systems, 1, 77-85. http://dx.doi.org/10.1016/j.hlpt.2012.04.005
- Peixoto, H., Duarte, J., Abelha, A., Santos, M. and Machado, J. (2012) ScheduleIT-Open-Source Preventive Actions Management Plataform in Healthcare Information Systems. Procedia Technology, 5, 734-742. http://dx.doi.org/10.1016/j.protcy.2012.09.081
- TRC Trisomy Risk Calculator. http://www.sbpsoftware.com/es/trisomy-risk-calculator.html
- Roche (2014) Essentials in Down Syndrome Screening. http://www.roche-diagnostics.ch/content/dam/corporate/roche-dia_ch/documents/broschueren/professional_diagnostics/immunologie/praeeklampsie/05109361001_EN_EA_Down-syndrome-screening_Brosch%C3%BCre_EN.pdf
- Kraus, C. (2014) Inside Goal-Directed Design: A Two-Part Conversation with Alan Cooper. http://www.cooper.com/journal/2014/04/inside-goal-directed-design-a-two-part-conversation-with-alan-cooper
- Palomaki, G.E. and Haddow, J.E. (1987) Maternal Serum α-Fetoprotein, Age, and Down Syndrome Risk. American Journal of Obstetrics & Gynecology, 156, 460-463. http://dx.doi.org/10.1016/0002-9378(87)90309-7
- Cuckle, H., Wald, N. and Thompson, S. (1988) Estimating a Woman’s Risk of Having a Pregnancy Associated with Down’s Syndrome Using Her Age and Serum Alphafetoprotein Level. International Journal of Gynecology and Obstetrics, 26, 336. http://dx.doi.org/10.1016/0020-7292(88)90310-4
- Reynolds, T.M. and Penney, M.D. (1990) The Mathematical Basis of Multivariate Risk Screening: With Special Reference to Screening for Down’s Syndrome Associated Pregnancy. Annals of Clinical Biochemistry, 27, 452-458.
- ROCHE (2011) Current Trends in Antenatal Screening for Down’s Syndrome. 175-177.
- Yaron, Y., Cherry, M., Kramer, R.L., O’Brien, J.E., Hallak, M., Johnson, M.P. and Evans, M.I. (1999) Second-Tri- mester Maternal Serum Marker Screening: Maternal Serum α-Fetoprotein, β-Human Chorionic Gonadotropin, Estriol, and Their Various Combinations as Predictors of Pregnancy Outcome. American Journal of Obstetrics & Gynecology, 181, 968-974. http://dx.doi.org/10.1016/S0002-9378(99)70334-0
- Wald, N.J., Cuckle, H.S., Densem, J.W., Nanchahal, K., Royston, P., Chard, T., Haddow, J.E., Knight, G.J., Palomaki, G.E. and Canick, J.A. (1988) Maternal Serun Screening for Down’s Syndrome in Early Pregnancy. Vol. 297, 883-887.
- Benn, P.A., Clive, J.M. and Collins, R. (1997) Medians for Second-Trimester Maternal Serum α-Fetoprotein, Human Chorionic Gonadotropin, and Unconjugated Estriol; Differences between Races or Ethnic Groups. Clinical Chemistry, 43, 333-337.
- Soft, S.B.P. and Maragall, J. (2007) Programapara la detección de la trisomía 21.
- Digital, G.O. (2007) No. 25908 Gaceta Oficial Digital, lunes 29 de octubre de 2007 1. 1-2.
- Cuenca, C., Gomez, J. and Scotti, S. (2012) Design an Architecture That Allows the Interoperability between Information Systems, Software or Medical Device Adopting the Standard.
- Barbarito, F., Pinciroli, F., Mason, J., Marceglia, S., Mazzola, L. and Bonacina, S. (2012) Implementing Standards for the Interoperability among Healthcare Providers in the Public Regionalized Healthcare Information System of the Lombardy Region. Journal of Biomedical Informatics, 45, 736-745. http://dx.doi.org/10.1016/j.jbi.2012.01.006
- Roncancio, G.E. (2009) Ayudasdesde la red para el control de la epidemia. Infectio, 13, 217-222. http://dx.doi.org/10.1016/S0123-9392(09)70152-3