Open Journal of Genetics
Vol.1 No.2(2011), Article ID:7314,3 pages DOI:10.4236/ojgen.2011.12004
Consequences of primer binding-sites polymorphisms on genotyping practice
![]()
1Institute of Molecular Pathology and Immunology of the University of Porto (IPATIMUP), Porto, Portugal;
2Faculty of Sciences, The University of Porto, Porto, Portugal;
3Medical Genetics Center, National Institute of Health, INSA, Porto, Portugal;
4University of Trás-os-Montes and Alto Douro, Institute for Biotechnology and Bioengineering, Centre of Genomics and Biotechnology, Vila Real, Portugal.
Email: *emartins@ipatimup.pt; lazevedo@ipatimup.pt; aamorim@ipatimup.pt; laura.vilarinho@insa.min-saude.pt; asofiaesteves@gmail.com; monicaslm@hotmail.com
Received 11 May 2011; revised 29 June 2011; accepted 10 July 2011.
Keywords: Primer Binding-site; SNP; PAH Gene; Molecular Genetics
ABSTRACT
Herein we investigated the effect of primer binding site polymorphisms in achieving correct genotyping when a mismatch occurs in distinct positions of the primer sequence. For that purpose primer sequences were designed in order to carry either allelic form at the 3’ end and at 3 bp, 5 bp and 7 bp apart from the 3’end of an intronic polymorphism (rs2247836) observed in phenylalanine hydroxylase (PAH) gene. For one of the alleles annealing failure was obtained when the mismatch occurs at all the four primer-site locations. Primer sequences carrying the alternative SNP allele resulted to be less specific as the distance to the primer-3’ end was increased. Altogether, these results revealthat effects in the extension of the annealing failure is allele and mismatch-position dependent.
1. INTRODUCTION
A crucial step in molecular genotyping is the primer design. Ideally, primers should match “cold” regions of genomic variability in order to avoid allelic drop-outs due to annealing failure in binding sites [1,2] . Cases reporting genotyping inconsistencies due to primer-site mismatches have been mainly documented in forensics [3] but there are also cases reported in genetic testing genotyping [4,5] . Any primer sequence carrying one of the two alleles of a polymorphic site is expected to result in genotype inconsistencies. When the mismatch occurs during the amplification of a homozygous sample, a null allele is thus expected (lack of amplification product)whereas in heterozygous individual for the primer binding site SNP a single chromosomes is amplified, and finally, in homozygous individuals for the other allele where no annealing failure occurs the genotype is correctly recognized. Hence, the extension of the genotype failure would be strongly dependent on the allelic frequencies of any SNP embedded at a primer binding site.
Another interesting issue is the position of the mismatched nucleotide within the primer sequence and the diversity observed in the final sequencing results. Expectedly, mismatches at the 3’end or close to the 3’end of the primer sequence would likely result in annealing failure but data is still scarce in what concerns mismatches at other positions of primer sequences. Although we are aware that this issue depends on aspects such as, for instances, the primer sequence, the amplification temperature and the sequence of the target DNA, a more detailed investigation would provide useful information. In this perspective, we used the phenylalanine hydroxylase gene (PAH) to understand how mismatches at distinct primer sites would affect the accuracy of the resulting genotypes.
2. MATERIAL AND METHODS
2.1. Marker Selection
An intronic SNP of the PAH gene (rs2247836; alleles A/G) located 167bp upstreamexon 5 was selected based on allelic frequencies available at the HapMap project [6] (Figures 1(a), (b)). Primer sequences were designed to match each of the SNP alleles exactly at the 3’end (P1), 3 (P3), 5 (P5) and 7 (P7) nucleotides from the 3’end, respectively (Figure 1(c)). The common reverse primer used was 5’CCACACACACATACACGCATGC3’.
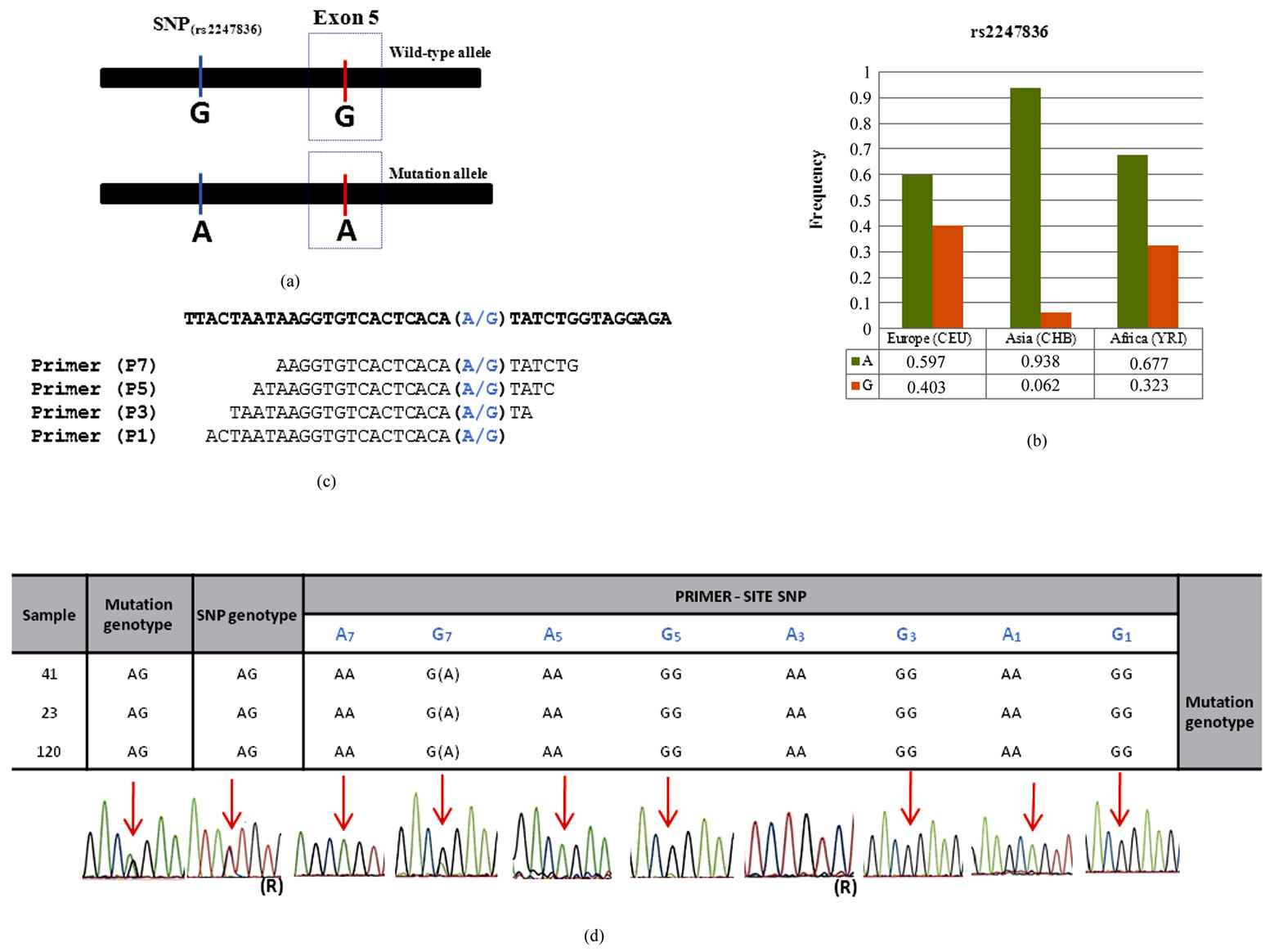
Figure 1. (a) Linkage disequilibrium between the intronic rs2247836 and the Arg158Gln mutation site; (b) Allelic frequencies of the rs2247836 in European (CEU), Asian (CHB) and African populations (YRI) as retrieved from the HapMap project [6]; (c) Primer sequences showing each of the rs2247836 alleles in blue; (d) Results obtained with P1, P3, P5 e P7 primers ((R) means the amplification with reverse primer).
2.2. SNP Amplification and Detection
PCR amplifications were performed in a 10 µL total reaction volume comprising QIAGEN PCR Master Mix, 0.2 mM reverse and forward primers and 5 - 10 ng DNA under the following conditions: 95˚C for 15 min of initial denaturation and 35 cycles of 94˚C for 30 s, 60˚C for 30 s and 72˚C for 1 min and 30 s, and a final extension at 72˚C for 10 min. The samples were purified through Sephadex G-50 columns and analyzed with an ABI PRISM 3130 Genetic Analyzer. The heterozygous status concerning the rs2247836 SNP and the PAH-Arg158Gln mutation was established using an external forward primer (5’GCTGTAGATGAGGTTTCTTTAAGAAC3’).
3. RESULTS AND DISCUSSION
Our strategy was based in the identification of an intronic SNPs in the neighborhood of the PAH exon 5 where a primer sequence could be designed to assess the accuracy of the previously identified genotype of three heterozygous individuals for the Arg158Gln mutation, referred to one of our center to PAH diagnosis (MIM 612349). Since the most closely linked polymorphic sites upstream of exon 5 (rs3817446 and rs1568791) have no frequency data available, we targeted rs2247836 (A/G) located 167 bp upstream exon 5 (Figure 1(a)) and with allelic frequencies ranging from approximately 60% for the A allele in Europe to 94% in Asia (Figure 1(b)) [6] .The three samples here analyzed are thus heterozygous for both the polymorphic site and the mutation site.
The results (Figure 1(d)) show specific amplification of the chromosome for which a primer matching sequence was used in three SNP-site locations: P1, P3 and P5, although in P5 carrying the G allele (G5) a week signal was obtained for the non-specific SNP-A allele. Lack of specificity was obvious with P7 carrying the G allele (G7) revealing a strong signal for the non-specific SNP-A allele.
Remarkably, amplification with the A7 primer resulted in a specific amplicon. That is, a primer carrying the A allele at the 3’end or at 3, 5 and 7 nucleotides from the 3’end would result in specific amplification of the A allele and failure in detecting SNP-G chromosomes and all the remaining variation in linkage disequilibrium is therefore expected to be lost.
4. CONCLUSIONS
In summary, we present a case study showing the effect that distinct alleles and distinct mismatching locations may have in genotyping practice, specifically in defining the homozygous/heterozous status of a closely linked allele. As the number of SNPs is increasing at the public databases we hope this information to be useful to all those involved in genotyping practices, namely in molecular analyses.
5. ACKNOWLEDGEMENTS
This work was supported by the Fundação para a Ciência e a Tecnologia (FCT) grant (PTDC/SAU-ESA/72663/2006) and by POPH-QRENTipologia 4.2-Scientific employment promotion funded by the European Social Fund and by national funds from the Ministry of Science, Technology and Higher Education (MCTES) to L.A. The Institute of Molecular Pathology and Immunology of the University of Porto (IPATIMUP) is an Associate Laboratory of the Portuguese MCTES and is partially supported by FCT.
REFERENCES
- Koboldt, D.C., Miller, R.D. and Kwok, P.Y. (2006) Distribution of human SNPs and its effect on highthroughput genotyping. Human Mutation, 27, 249-254. doi:10.1002/humu.20286
- Quinlan, A.R. and Marth, G.T. (2007) Primer-site SNPs mask mutations. Nature Methods, 4, 192. doi:10.1038/nmeth0307-192
- Alves, C., et al., (2001) VWA STR genotyping: Further inconsistencies between Perkin-Elmer and Promega kits. International Journal of Legal Medicine, 115, 97-99. doi:10.1007/s004140100215
- Wong, L.J., et al. (2001) Novel SNP at the common primer site of exon IIIa of FGFR2 gene causes error in molecular diagnosis of craniosynostosis syndrome. American Journal of Medical Genetics, 102, 282-285. doi:10.1002/ajmg.1461
- Jeffrey, G.P., et al. (1999) Polymorphism in intron 4 of HFE may cause overestimation of C282Y homozygote prevalence in haemochromatosis. Nature Genetics, 22, 325-326. doi:10.1038/11892
- Frazer, K.A., et al. (2007) A second generation human haplotype map of over 3.1 million SNPs. Nature, 449, 851-861. doi:10.1038/nature06258

