Advances in Internet of Things
Vol.4 No.3(2014), Article ID:47538,9 pages
DOI:10.4236/ait.2014.43004
LOD Cloud Mining for Prognosis Model (Case Study: Native App for Drug Recommender System)
Nidhi Kushwaha*, Raman Goyal, Pramiti Goel, Sidharth Singla, Om Prakash Vyas
Department of Information Technology, Indian Institute of Information Technology (IIIT-A), Allahabad, India
Email: *Kushwaha.nidhi12@gmail.com, raman.goyal111@gmail.com, pramitigoel20@gmail.com, sidpkl.singla@gmail.com, opvyas@iiita.ac.in
Copyright © 2014 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 30 April 2014; revised 30 May 2014; accepted 30 June 2014
ABSTRACT
The goal of this project is to use the Semantic Web Technologies and Data Mining for disease diagnosis to assist health care professionals regarding the possible medication and drug to prescribe (Drug recommendation) according to the features of the patient. Numerous Decision Support Systems (DSS) and Expert Systems allow medical collaboration, like in the differential diagnosis specific or general. But, a medical recommendation system using both Semantic Web technologies and Data mining has not yet been developed which initiated this work. However, it should be mentioned that there are several system references about medicine or active ingredient interactions, but their final goal is not the Drug recommendation which uses above technologies. With this project we try to provide an assistant to the doctor for better recommendations. The patient will also able to use this system for explanation of drugs, food interaction and side effects of corresponding drugs.
Keywords:Linked Life Drug Data Cloud, Recommender System, Data Mining, Semantic Web

1. Introduction
Drug overdosing or under dosing, or the consumption of any drug together with other drugs or foods, may result in undesirable outcomes of Adverse Drug Events (ADEs), which may be life threatening and even lead to death [1] . More than one lakhs injuries are held because of ADEs, the survey of (AHRQ), which causes death and extra expenditure to the hospital every year [1] [2] . Though, from 28% to 95% of ADEs could be prevented by reducing medication errors. Today, there are many useful drugs and ADE related knowledge sources available on the web that can be utilized to discover existing or even new potential ADEs and provide timely alerts to relevant patients [3] . Some examples for that are US Food & Drug Administration (FDA), Med-Watch alerting service, online medical digital libraries such as PubMed ADE (ontological knowledge sources) such as SIDER [4] and Drug Bank [5] . It provides access to various Knowledge Bases for mutual extraction of informative data. Other drug related websites such as Drugs.com and Medline Plus also provide valuable information about various drugs, potential interactions, and side-effects, via simple keyword based searches. In spite of the luxuriant amounts of knowledge about Drugs and ADEs, it still remains a profound challenge for patients or even their physicians to discover relevant ADE knowledge. The “information barriers” that users face can be classified into three main types. First, users may be unaware of the existence of these sources. Second, many users, especially patients, usually who are not familiar with the Drug and ADE terminology. Medical Professional users usually lack of the required technical skills for correctly expressing their information needed for accessing ADE knowledge, which is provided by Drug Bank and SIDER. Third, Identification of Drug-Drug and Drug-diseases, interactions are also needed before recommending. Extracting these is a very difficult problem, because of increment of the large number of available drugs coupled with the ongoing research activities in the pharmaceutical domain. Although some international standards like ICD-10 [6] classification and the UNII registration need to be continuously updated before taking final decision using them. Using the real time data extracted from various medical camps and expanding it, which includes personal information about the patients, such as the list of medications the patient is currently prescribed by, we can recommend drugs for a patient having another set of diseases while trying to minimize the risk of Drug-Drug interactions and drug side effects. The so called “marriage” [7] [8] of the technologies brings an opportunity to use it with a mobile application. We are blessed with the logically related open information [1] -[3] [9] [10] which is available online. Our proposed work utilizes this information for the recommendation. The goal of the project is to use the Semantic Web Technologies and Data Mining for disease diagnosis to assist health care professional regarding the possible medication and drug to prescribe (Drug recommendation) according to a criteria. Numerous Decision Support Systems (DSS) and Expert Systems allow medical collaboration, like in the Differential Diagnosis specific or general. A medical recommendation system using both Semantic Web technologies and Data Mining has not yet been developed which initiated this project work. However, it should be mentioned that there are several system references about medicine or active ingredient interactions, but their final goal is not the Drug recommendation which uses above technologies.
2. Related Work
Semantic Web technologies [11] can be efficiently accessed through the advancement in the field of their memory efficient storage and easy fetching through code [12] [13] [14] . This knowledge, reasoning can be utilized for data classification, understanding the medical records. Different recommendation approaches have been developed using a variety of methods. Semantic recommender systems form a class of Recommender systems that make use of ontology’s, context awareness, and other semantic methods to make informed recommendations. For example, some researcher Middleton et al. [7] [15] argue for an ontological approach to user profiling. They do this by monitoring user behavior in the selection of recommended academic research papers, and coupling that with relevance feedback. Semantic recommendation systems are characterized by the incorporation of semantic knowledge in their processes in order to improve recommendation’s quality. In 2005, Adomavicius and Tuzhilin [16] published a survey of recommender systems, which classified them into three main approaches— content-based, collaborative, and hybrid systems. Further, they examine the recommended techniques used for each approach. The authors argue for extending the capabilities of extant systems by providing: 1) improved models of users and “items”; 2) incorporation of the contextual information into the recommendation process; support for multi-criteria ratings; and the provision of a more flexible and less intrusive recommendation process. More specifically, with the use of Ontology languages such as OWL, a rather large amount of biomedical Ontologies have been developed among them (BioPax) [17] , the GALEN ontology [18] and the Foundational Model of Anatomy (FMA) etc. Some of the research projects funded for it are TUMOR, REMINE and PSIP [3] [19] -[20] . REMINE project is to build a high performance prediction, detection and monitoring platform for managing Risks against Patient Safety (RAPS). PHE [20] introduces the concept, design, and implementation of the Personal Health Explorer (PHE). This system provides semantically enhanced recommendations that can be stored in the individual’s PHR for further research and consultation. NeOn [9] and Active Semantic Documents [20] uses Ontologies [20] in the daily routine medical treatment. In our work we have used semantic data for feature generation. The idea of feature generation from LOD Cloud has been previously discussed by Heiko et al. in the paper [10] [21] .
3. Proposed Work
Nowadays, new dedicated IT solutions for the patient safety [22] domain are continuously being explored, aiming to help people to prevent mistakes by medical teams. With this project we aim to develop an approach (Figure 1, Block Diagram and Figure 2, Detailed Flow Diagram) that helps to increase patient safety by helping health care professionals to relevant information that is gathered from database obtained from various Semantic Web sources educating them toward better preventive medicine decision making. We find a patient can be suffering from multiple diseases and can be prescribed different medicines for their respective diseases by health care professionals. However, the medicine prescribed for one disease might have adverse effects when it would be consumed along with other curable drugs. In this project, we present a web based system which not only suggests the user about which drug is to be taken, but also to avoid adverse effects of Drug-Drug interaction and Drug-Allergy interaction. Our drug recommender system features three functionalities:
1) Recommendation of the best drug for the given set of Inputs.
2) Using Semantic knowledge of LODD cloud.
3) Depending on the severity of ADE, warns of possible adverse effects the conflicting the medicines might have. Keep a complete history of patient’s drugs past intake with its effect.
Our experimental result will show that the use of semantic descriptions of information items combined with
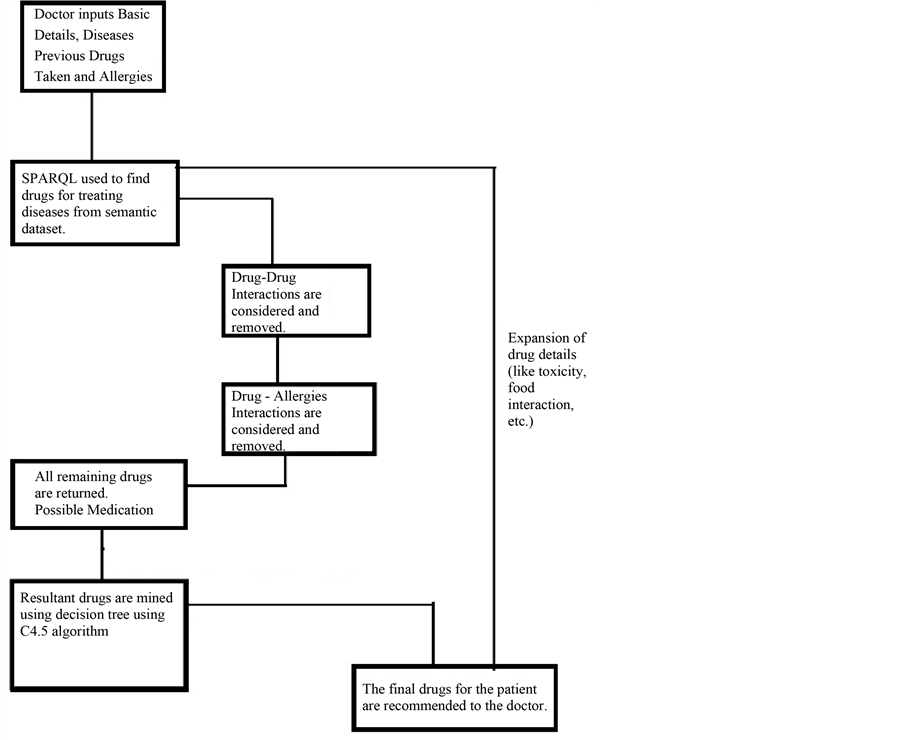
Figure 1. Block diagram of proposed work.
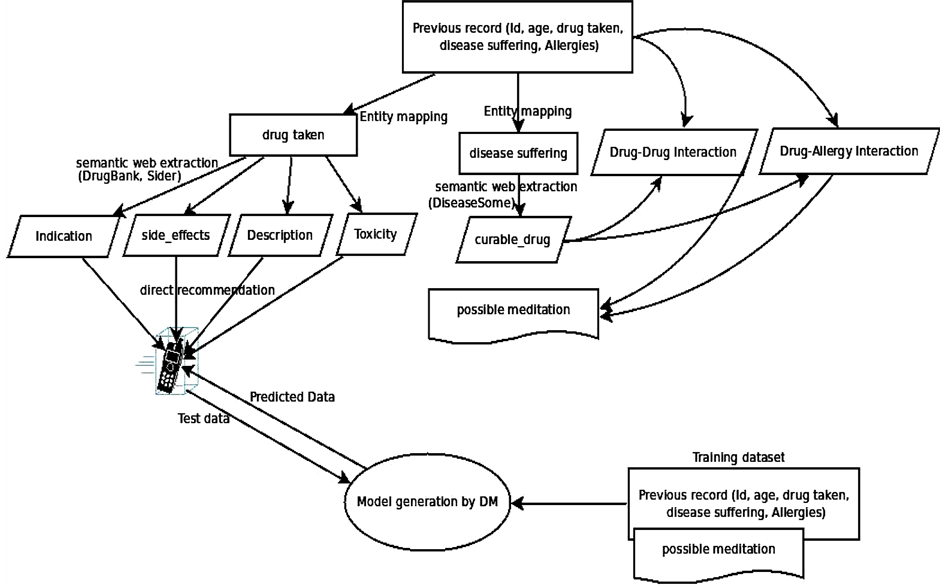
Figure 2. Detailed Flow diagram.
the multi-attribute features improve the accuracy of the suggestions and the quality of recommendations. The use of Semantic Web technologies has been found to be a good match for developing Drug recommendation systems. Ontologies can effectively encapsulate medical knowledge and rule-based reasoning can capture and encode the drug interactions knowledge. We are going to recommend suitable drug for the user which is not a layman to understand its suitability but a more sophisticated one. We give the recommendation with the help of Data Mining [23] & Semantic Technologies.
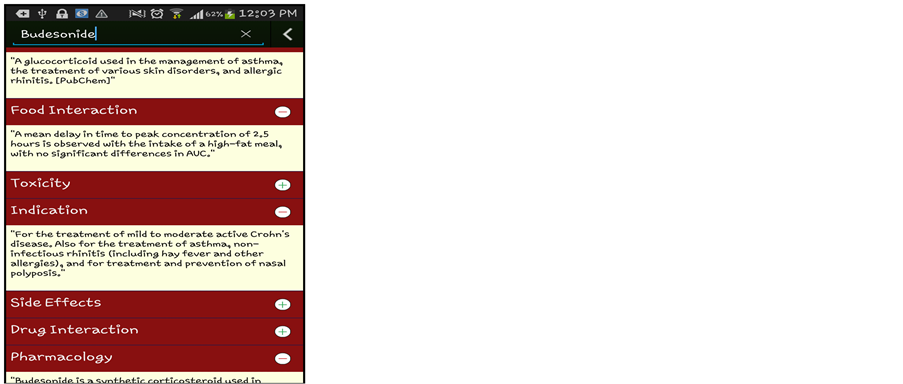
Simple Data Mining algorithms can’t able to consider the domain specific information, in our work, this information will boosted with the combination of Semantic Web and Data Mining. This semantic information is obtained from the Linked Open Life Science Data (the semantically linked information related to drugs, its Side-effects and Allergies). After the input process is completed feature generation process gets initiated which fetch related information from the cloud with the help of SPARQL querying Table1 In table F1, F2, F3…..F6 are the attributes (age, gender, BMI, drugs taken, disease suffering, allergies) of the patient. While the attributes F7,….,F9 will be generated by attribute F4 (drug taken) and F5 (disease suffering). As we can see from the table that attributes can be multivalued. So, the generated attribute consists of the information like, in the first row attribute F5 (disease suffering) has two diseases A, B. The attributes generated by A and B will be treated as independent and named as A_curabledrug,..A_toxicity, B_curabledrug….B_toxicity. For simplicity, we can say that from attribute F7 (generated attribute) we retrieved the information about its (drug) ID from the Diseasesome (RDF data) and explore it further to obtain the curable disease, drug’s side effects and its food interaction, etc. Some features are manually chosen for directly represent in the final output screen like: food interaction, toxicity, description, indication and side effects (see Figure 3) while other features (like: drugs, allergy) are used for generating a suitable pair of drugs.
The output of this step serves as the input of next two processes; one is a Drug-Drug interaction (see Section 6) and the other is for final GUI (see Figure 4). Drug-Allergies are obtained by considering all the remaining Drugs that helped for treatment. Last step, is to combine the result of Drug-Drug and Drug-Allergy interaction with the historical data and then proceed for mining. Last but not the least is to make a final recommendation with the interactive app so that it can easily view by the patient and (mainly for) doctor. In the next section authors explained about how to combine the Semantic Web queries and data mining algorithm.
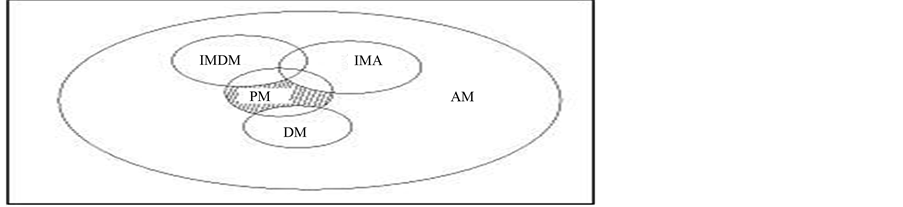
Figure 3. Drug-drug interactions.
4. Combine Mining & Semantic Queries
Data mining techniques usually directly apply to actual data records. But, it was challenging to combine them with semantic data and then apply mining algorithm to it. For that author first store the RDF data into storage then retrieved this information to enrich the features as described in the previous section. Initially we have patient records with some features like patient id, disease, previous Allergies, previous drug etc. For enhancement we choose disease name for generating disease id and related information from the Semantic Web cloud. Basically, this task is known as Feature Generation. From there we also get the information about various features like to relate drugs, drug toxicity, food interaction, disease indications (see Figure 1). For recommendation with both semantic and mining technologies, the features generated by the semantic technology are explicitly pruned with the help of manual revision. Here, manual revision has been done by selecting some semantically generated features manually for Data Mining task, while on the other side, the rest of the features are directly fed to the final user’s GUI. Some features for ex: Disease suffered, Curable Drug along with Age, Sex, Smoking/NonSmoking and Drinking/NonDrinking as their class label are mine for the recommendation of new cases. On the other hand, other features like Food interaction, description, toxicity and side-effects are directly fed to the end user’s GUI (in Figure 2 shown by “Expansion of drug details” link). These separations of features have been done by manual revision as explained above. This system is mainly developed for professionals or Doctor’s while at the same time it will be informative to the layman or patients also. For example: Recommended drug pairs is good for a Doctor after entering all details of the patient’s while Food interaction, drug description, toxicity and side-effects will be useful of the patient for more understanding about his/her disease as well as drug.
Data Sets Description: In the mining phase, we also consider the other attributes present in the database like BMI, Smoking/Non Smoking, Age, and Drinking/Not Drinking in combination of disease, previous drugs and their class label.
5. Detail Explanations (Drug-Drug Interaction)
For the particular diseases LODD cloud will return drug pairs. Our aim is to find the drugs that do not interact with the previous drug taken by the user and the proposed drug pair from the LODD cloud. So, the authors reduce the pair up to the position so that the final solution is the smallest group obtained from bigger groups. Here, in Figure 3 AM represent all the drugs which stored in the ontology and over which reasoning can be performed. DM is the drugs associated with the patient and therefore it should be excluded from final recommendation. Next, IMDM represents drugs that interact with the currently prescribed to the patient (those in DM Group).


 (a)
(a)


 (b) (A)
(b) (A)

 (B)
(B)
Figure 4. (A) Sample of Drug Recommendation System (for Health Care Professionals); (a) Drug Recommendation for Patient 1; (b) Drug Recommendation for Patient 2; (B) Sample of Drug Recommendation System (for Layman & Health Care Professionals).
IMA, drugs interact with the patient’s allergies. Lastly the PM, are those groups which we are interested and can be recommended to the patient.
Example Explanation:
Suppose the following parameters as an example (input parameters):
1) Basic Information (Ram, 18, M)
2) Disease: D 3) Previous Drugs:{MA, MB, MC}
4) Allergies: AL1 With the following information the system is capable to obtain a drug list that could heal a given disease “D” without any drug-drug interaction. Here, DM= {MA, MB, MC} is the drugs that will be excluded because they are already used. Drugs that interact with the previously taken drugs are also excluded and we call it IMDM = {MD, ME, MF, MH, ML}. To avoid patient’s allergies interaction, IMA = {MM, MN, MO} also gets excluded. List of possible medications for patient recommendation is PM = {MP, MQ, MV}. The system must divide the PM group into n subgroups. Every subgroup must represent a type of medicine (anti pain, anti infection, etc.) and all medicines contained in each group must be of the same type. The system will return these pairs (drug tuple) for the expert (Data Mining) to recommend the patient one of those pairs. This prediction is based on the whole real dataset of the patient (BMI, Gender, Smoking/Nonsmoking, and Age) including the features from LODD and drug tuples as the class label. Finally the drug tuple is recommended by the doctor for final decision. Updation in training is also possible by appending the test data with the predicted value.
6. Results & Discussion
Semantic Web has structured web documents where drug and its information are interlinked. Linked Open Drug Data (LODD) [6] project facilitate this integration by bringing these medical data sources onto the Web of Linked Data. This shows the combination of two major technologies which results in Semantic Web Mining because simple Data Mining cannot consider the domain specific information. We are boosting our recommendation system with the domain specific information which is coming from the Linked Open Drug Data (the semantically linked information related to various domains). Here, we present this technology for Drug Recommender Systems but this can be easily applied to other domains as well. The procedure involves retrieving information from LODD and including possible Drug-Drug interaction. The database (RDF dump stored in Sesame) can be updated as needed. The storage provides the update option through which we can update our stored information. So, the stored information is not static, it can be changed as new RDF triples are available for use. After enrichment with the semantic knowledge this can be mined to predict previously unseen information. The results are mined with data mining C4.5 algorithm and bagging [23] , displayed in Figure 5. The proposed system will be used by the healthcare professionals as they know medical terms of the drugs. At the same time semantic knowledge provides some more information for layman ex. Side effects, food interactions and description of the Drugs. Without semantic enrichment, simple mining on the health record are dependent only on a limited number of features present in that current dataset. This information surely predicts no extra advantage in prediction result [23] . So we move forward for a budding technology, Semantic Web and incorporate into our system before applying DM techniques. The structured drug information already present in open source form called LODD. Relatedness of the drug and disease in the logical form provide an opportunity for extracting more meaningful features like toxicity, food interaction, etc. When we use semantics of LODD and filteration, we get
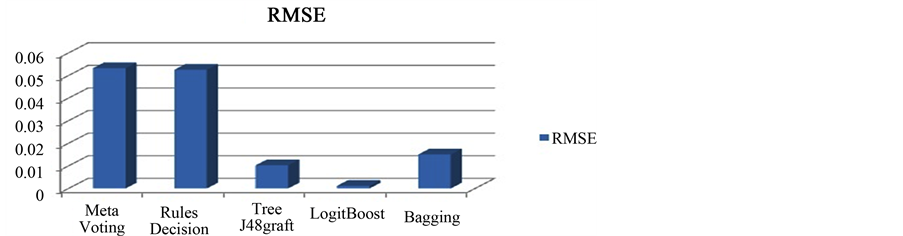
Figure 5. RMSE b/w 5 Data Mining algorithms.
only Drug-Allergy and Drug-Drug interactions. However, mining technologies help in further improving results by taking other important attributes like age, BMI, Smoking and Drinking habits into account because these factors highly influence recommendation of the Drug-set for the patient. If new disease or new Drug (Not present in the previous data) is inserted by the doctor, then mining will not predict any result. But, Semantic Querying will extract that information from LODD. So we can say that Mining and SW provide complementary results. In Figure 4(A) & Figure 4(B)) authors have shown different pictures of a developed Drug Recommender System (DRS) for two patients. For the comparison of different data mining algorithms we have taken some of the algorithms into our consideration and perform analysis (in Figure 5) with the help of Weka data mining tool [24] . It shows ensemble learning techniques are more suitable for the classification task. For the app implementation, we follow the process model of mConcAppt [25] concept of Mobile Software Engg. The App uses the server of Sesame [26] where all the data already been stored. Ontologies Used: We have downloaded the dump of SIDER (for side effects) [4] , Drug Bank [5] into the Sesame RDF repository and querying it with the help of SPARQL 1.1 with Java IDE.
7. Original Contributions
Summarizing the aforementioned discussions we conclude following points:
We have done Drug-Drug and Drug-Allergy interaction using semantic web knowledge. After this we get a pair of drugs that are suitable for a particular disease for a patient having some allergies and was taking drugs before prescription from our method.
Prediction of drug pairs can be seen by two subparts below.
1) If the training data already have similar information related to test data;
We consider these drug pairs as a class label and other attributes like age, gender, BMI, drugs taken, having allergies as a feature, then we predict the drug pair of the people who have the same disease, same allergies and have taken same drugs previously.
2) If training data have no similar information related to test data;
In this case we get Drug-Drug pair from SW, but DM will not produce any result. So, for this case we add this new sample (containing BMI, age, gender, drug taken, having allergies and lastly Drug-Drug pair) to training data so that next time our training data will also predict the similar kind of information that was unknown to it previously.
By former two points we combined both SW and DM techniques for assisting (mainly to) health care professionals as well as the patient.
References
- (2014) Sustainment Guide for Adverse Drug Events, Partnership for Patients Compaingn, Healthcare Government.
- Chen, B., Ding, Y. and Wang, H. (2010) Chem2Bio2RDF: A Linked Open Data Portal for Systems Chemical Biology. Proceedings of 2010 IEEE/WIC/ACM International Conference on Web Intelligence and Intelligent Agent Technology (WIIAT), Toronto, 31 August-3 September 2010, 232-239. http://dx.doi.org/10.1109/WI-IAT.2010.183
- Ceusters, W., Capolupo, M., Moor, D. and Devlies, J. (2008) Introducing Realist Ontology for the Representation of Adverse Events. Proceedings of the 2008 Conference on Formal Ontology in Information Systems (FOIS 2008), 237-250.
- Jentzsch, A. (2013) Sider. http://datahub.io/dataset/fu-berlin-sider/resource/e84dd6f3-f22e-4d4d-9ee8-e5b004eb654c
- Jentzsch, A. (2013) Drugbank. http://datahub.io/dataset/fu-berlin-Drugbank/resource/8fc23108-81d0-45f2-81ec-22ea41485f49
- Williams, W. and Mancini, L. (2011) ICD-10 White Paper: Data Impact Across the Enterprise. The Kiran Consortium Group LLC, 1-7.
- Sheth, A. (2005) Semantic Web & Semantic Web Services: Applications in Healthcare and Scientific Research. Proceedings of IFIP Working Conference on Industrial Applications of Semantic Web, 188.
- Morrell, T. and Kerschberg, L. (2012) Personal Health Explorer: A Semantic Health Recommendation System. IEEE 28th International Conference on Data Engineering Workshops (ICDEW), Arlington, 1-5 April 2012, 55-59. http://dx.doi.org/10.1109/ICDEW.2012.64
- Gómez-Pérez, A. and Suárez-Figueroa, M.C. (2009) NeOn Methodology for Building Ontology Networks: A Scenario-Based Methodology. Proceedings of the International Conference on Software, Services & Semantic Technology, Sofia, 28-29 October 2009.
- Paulheim, H. and Furnkranz, J. (2012) Unsupervised Generation of Data Mining Features from Linked Open Data. Proceedings of Web Intelligence, Mining and Semantics, WIMS’12, Craiova, 13-15 June 2012. http://dx.doi.org/10.1145/2254129.2254168
- Bizer, C., Heath, T. and Lee, T.B. (2009) Linked Data—The Story So Far. International Journals Semantic Web Information System, 3, 1-22.
- Prudhommeaux, E. and Seaborne, A. (2008) Sparql Query Language for RDF, W3C Recommendation.
- Marshall, M.S., Boyce, R. and Deus, H.F., et al. (2012) Emerging Practices for Mapping and Linking Life Sciences Data Using RDF—A Case Series. Web Semantics: Science, Services and Agents on the World Wide Web, 14, 2-13. http://dx.doi.org/10.1016/j.websem.2012.02.003
- Bauer, F. and Kaltenbock, M. (2011) Linked Open Data: The Essential. Proceedings of Finite Element Analysis and CAD, Peking University Press, Beijing, 9-15.
- Middleton, E., Shadbolt, N. and Shadbolt, D.C. (2004) Ontological User Profiling in Recommender Systems. Proceeding of ACM Transactions on Information Systems (TOIS), 22, 54-88.
- Adomavicius, G. and Tuzhilin, A. (2005) Toward the Next Generation of Recommender Systems: A Survey of the State-of-the-Art and Possible Extensions. Proceedings of IEEE Transactions on Knowledge and Data Engineering, 17, 734-749. http://dx.doi.org/10.1109/TKDE.2005.99
- Wolstencroft, K., Brass, A., Horrocks, I., Lord, P.W., Salter, U., Turi, D. and Stevens, R. (2005) A Little Semantic Web Goes a Long Way in Biology. Proceedings of ISWC2005, 786-800.
- Rector, A. and Rogers, J. (2006) Ontological and Practical Issues in Using a Description Logic to Represent Medical Concept Systems: Experience from GALEN, Reasoning Web, Second Int Summer School, Tutorial Lecture 06, 4126, 197-231.
- Ruttenberg, A., Rees, J. and Luciano, J. (2005) Experience Using OWL DL for the Exchange of Biological Pathway Information. Proceedings of the 1st OWL Experiences and Directions Workshop, Galway, 11-12 November 2005, 1-12.
- Aranguren, M., Fernandez-Breis, J. and Dumontier, M. (2013) Special Issue on Linked Data for Health Care and the Life Science. Department of Biology, Carleton University, iOS Press.
- Paulheim, H. (2012) Explain-a-LOD: Using Linked Open Data for Interpreting Statistics. In: Duarte, C., Carrico, L., Jorge, J.A., Oviatt, S.L. and Goncalves, D., Eds., IUI’12 Proceedings of the 2012 ACM International Conference on Intelligent User Interfaces, ACM, New York, 313-314.
- Beuscart, R., McNair, P. and Brender, J. (2009) Patient Safety through Intelligent Procedures in Education: The PSIP Project. Studies in Health Technology and Informatics, 148, 6-13.
- Han, J. and Kamber, M. (2000) Data Mining: Concepts and Techniques. Morgan Kaunn Publishers Inc., San Francisco.
- Hall, M., Frank, E., Holmes, G., Pfahringer, B., Reutemann, P. and Witten, I. (2009) The WEKA Data Mining Software: An Update, SIGKDD Explorations, 11.
- Hess, S., Kiefer, F., Carbon, R. and Maier, A. (2013) mConcAppt—A Method for the Conception of Mobile Business Applications. LNCS, Social Informatics and Telecommunications Engineering, Vol. 110, 1-20.
- Broekstra, J., Kampman, A. and Harmelen, F. (2002) Sesame: A Generic Architecture for Storing and Querying RDF and RDF Schema. ISWC, 2342, 54-68.
NOTES
![]()
*Corresponding author.


