Open Journal of Inorganic Chemistry
Vol.07 No.02(2017), Article ID:75871,33 pages
10.4236/ojic.2017.72003
Uranium Bio-Transformations: Chemical or Biological Processes?
Erica L.-W. Majumder, Judy D. Wall*
Department of Biochemistry, University of Missouri, Columbia, MO, USA

Copyright © 2017 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY 4.0).
http://creativecommons.org/licenses/by/4.0/



Received: February 28, 2017; Accepted: April 27, 2017; Published: April 30, 2017
ABSTRACT
Uranium bio-transformations are the many and varying types of interactions that microbes can have with uranium encountered in their environment. In this review, bio-transformations, including reduction, oxidation, respiration, sorption, mineralization, accumulation, precipitation, biomarkers, and sensors are defined and discussed. Consensus and divergences are noted in bioavailability, mechanism of uranium reduction, environment, metabolism and the type of organism. The breadth of organisms with characterized bio-trans formations is also cataloged and discussed. We further debate if uranium biotransformations provide bio-protection or bio-benefit to the microbe and highlight the need for more work in the field to understand if microbes use uranium reduction for energy gain and growth, as having the ability is separate from exercising it. The presentation centers on the fundamental drivers for these processes with an additional exposition of the essential contribution of inorganic chemistry techniques to the molecular characterization of these biological processes.
Keywords:
Uranium, Reduction, Bio-Sorption, Proteobacteria

1. Introduction
Metals are an essential part of life comprising cofactors, nutrients, chlorophyll, electron acceptors and donors, color creators and more. Although man has used metals extensively since copper was discovered in 9000 B.C.E., we are just beginning to appreciate the ability of biology to manipulate metals [1] . Uranium is not considered an essential metal for biological processes, but has become an essential part of lifestyle in the 20th century through the development of nuclear technologies from weapons to clean energy to medicine. Uranium is the largest stable element found on the planet Earth. While radioactive, the half-life is 4.5 billion years, meaning that it decays very slowly. Uranium is found naturally throughout the Earth’s crust with an average of 2.7 mg of uranium per kg of crust. Although uranium concentrations are much higher in naturally-occurring uranium deposits, some of which are mined for uranium [2] [3] , in the Earth’s crust, uranium is found mostly in mineral complexes [4] .
Despite the low-level natural abundance of uranium in soils, human activities have introduced dramatically high concentrations of uranium to water and soils near uranium mines, near other metal mines that also have high uranium content, near nuclear weapons manufacturing and testing sites, and near nuclear power plants―creating high concentrations of uranium in formerly pristine sites [5] . The vast majority of the human-introduced uranium has been a relatively recent activity. It started with the atomic age beginning in the 1930’s and escalated after the conclusion of World War II into the Cold War. Still today, more countries are attempting to make nuclear weapons or use nuclear power. These contaminated areas sometimes represent a significant global threat to human and environmental health and safety, demanding a need for remediation or immobilization by non-intrusive methods. Although the contaminating uranium is often depleted in the fissile 235U and, therefore, does not present a severe radiological risk, but 238U is still a highly toxic heavy metal [6] . Uranium poisoning affects humans by oxidatively damaging cells causing non-malignant respiratory disease and nephrotoxicity [7] . For the eco-systems at these sites, uranium pre- sents a significant challenge. Although uranium can be adsorbed to roots and be taken up by plants, processes that were initially considered possible remediation strategies, the toxic metal ultimately harms the plant [8] [9] . Similar damage occurs to microbes in that uranium causes oxidative damage to the cell and the DNA. However, despite the high toxicity and potential radiation damage from uranium, some naturally-occurring microbial communities have the ability to survive, and even thrive, in highly contaminated uranium conditions. These capacities yield the desired non-intrusive remediation or immobilization methods [10] [11] .
The ability to persist in a uranium contaminated environment is achieved through a number of different types of bio-transformations, typically chemical transformations of the metal carried out by a microbe. Microbes have been observed reducing, oxidizing, respiring, adsorbing, mineralizing, accumulating or precipitating uranium in the environment [12] . These interactions have been investigated as remediation strategies and in some cases characterized molecularly as unique chemical transformations and electron flow pathways. Microbes of all shapes and sizes have been found to have different interactions with uranium from Proteobacteria to fungi [13] [14] . The widespread nature of the bio- transformations of uranium both geographically and by various microbial families brings into question whether the bio-transformations are biologically or chemically driven? In other words, is any organism in the right redox environment capable of transforming uranium and are there biological advantages to the organisms carrying out these bio-transformations? An open question for researchers in the field has been whether these transformations are detoxification, resistance or just accidents of chemistry?
Previous reviews have discussed uranium reduction [12] , uranium bioremediation methods [13] [15] , uranium geochemistry, mineralization and groundwater transport [3] [16] . While all substantial and important reviews, the previous work has focused only on the most well-known organisms and do not consider the drivers of these processes or the biological benefit to the microbe beyond mentions of possible detoxification. In this review, we expand beyond the more commonly presented reduction and remediation to provide a current list of reported/characterized uranium bio-transformations as defined in Table 1 and schematized in Figure 1 and Figure 2 and to provide a broader but non-ex- haustive list of the organisms that carry out these chemical reactions in Table 2. In Table 3, we provide examples of the chemical reactions of representative uranium bio-transformations. We begin by examining the process of uranium reduction including the organisms, locations, mechanisms and rates. Then, we explore if uranium reduction is a form of cellular respiration or detoxification. In the second section we outline and define additional bio-transformations of uranium that include sorption, bio-markers and nanoparticle synthesis. We conclude by presenting the contributions of classical inorganic chemistry techniques to understanding uranium bio-transformations and by making suggestions about the chemical and biological drivers of this unique and surprising reaction between an actinide that is radioactive and toxic and many naturally-occurring microbes.
Table 1. Bio-transformations of uranium summary and definitions.
2. Bio-Transformations of Uranium
2.1. Uranium Reduction
2.1.1. Locations of and Organisms Capable of Uranium Reduction
Uranium reduction is the most mentioned form of microbial-based bio-trans- formation of uranium recorded in the literature. Uranium reduction is the process by which an organism chemically reduces uranium in the environment (or lab) from the VI oxidation state by a two electron process to the IV oxidation state. U(VI), the ionized species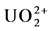 , is soluble in water while U(IV), unionized UO2, is insoluble (Table 3) [17] . This observation initially generated excitement as a potential bio-remediation or immobilization strategy. However, the reduced uranium could easily be reoxidized when environmental conditions changed and, therefore, simple reduction did not present a long-term remediation approach [18] [19] [20] [21] . Uranium reduction and geochemistry has been reviewed before [3] [12] [13] [16] so here we build on the recent studies.
, is soluble in water while U(IV), unionized UO2, is insoluble (Table 3) [17] . This observation initially generated excitement as a potential bio-remediation or immobilization strategy. However, the reduced uranium could easily be reoxidized when environmental conditions changed and, therefore, simple reduction did not present a long-term remediation approach [18] [19] [20] [21] . Uranium reduction and geochemistry has been reviewed before [3] [12] [13] [16] so here we build on the recent studies.
Uranium reduction is truly a global phenomenon and has been observed in multiple sites in the U.S. [10] , China [22] , France [23] , Germany [24] , South Africa [25] , U.K. [26] , Australia [2] and likely many others. Uranium reduction was originally thought to be possible only in anaerobic, reducing environments and carried out by known metal-reducing organisms [27] [28] . However, the perception of what organisms can reduce uranium has expanded considerably over the last two decades. Naturally, uranium reduction appears to occur at sites with high uranium concentrations, like acid-mine drainage or contaminated sediments. This contaminant has been shown to be a significant factor in microbial community formation creating communities that while less diverse, are more stable than communities in non-contaminated environments [29] . Uranium contamination drives distinct microbial community formation, but there are similar members when communities are compared globally [30] . The list of microbes now includes α, β, γ, and δ-Proteobacteria, Firmicutes, Actinobacteria and fungi and is detailed in Table 2. As an example, almost every class of microbes listed in Table 2 was found in the contaminated groundwater from the Oak Ridge Reservation, Oak Ridge, TN [9] . The first identified organisms capable of uranium reduction were already known metal reducers, primarily iron reducers. In the early 1990’s, microbial reduction of uranium was first reported for iron-reducing bacterial Geobacter species and followed closely by the sulfate-reducing bacterium Desulfovibrio desulfuricans [17] [31] . The third member of the “big three genera” in uranium reduction was Shewanella. Table 2 lists six Geobacteracae strains, six Desulfovibrionales, five Shewanella strains that have been identified to reduce uranium.
Other dissimilatory metal-reducing bacteria have also been shown to reduce uranium including Anaeromyxobacter dehalogens, which was found to tolerate higher oxygen concentrations and to associate with soil particles resulting in a different eco-physiology from the commonly seen uranium reducers [32] [33] . Diverging from the trend of Gram-negative Proteobacteria, strains of the Gram- positive spore-forming genus Desulfitobacterium reduce uranium to mononuclear
Table 2. List of organisms and their bio-transformations of uranium.
U(IV) not to the mineral uraninite [34] . Uranium reduction by other Gram- positive spore-forming Clostridium species was optimal in the pH of 4 - 6 with most being over pH 5 [35] . The authors of this review questioned if uranium reduction is a strictly anaerobic process because U(IV) exposure to air is readily converted to U(VI). However, aerobic organisms, a Bacillus [36] and a Caulobacter [37] , have also shown uranium reduction capacity. Additionally many of the well-known uranium reducers are facultative anaerobic organisms that live in suboxicconditions (Table 2), but a pyrosequencing study comparing uranium reducing communities in suboxic and anaerobic conditions identified more uranium reducing organisms in anaerobic conditions [38] . This suggests that the uranium reduction process proceeds more readily in anoxic conditions, but is possible in suboxic and oxic conditions.
The amount of oxygen in the environment is tied to the redox state. Iron reducers, sulfate reducers, nitrate reducers and other types of metal-reducing bacteria depend on an environment tuned to the redox potential of their metabolism in order to respire and gain energy. Sulfate reducers and iron reducers require a redox potential of about −200 mV, while nitrate reduction happens around +700 mV [39] . Uranium reduction requires reducing conditions, at least reducing enough―based on the electrochemistry or redox potential―to reduce U(VI) to U(IV). The reduction potential of the reaction is also dependent on the complexation of the metal, the pH of the solution and the concentration of ion. The redox potential has been measured in abiotic electrochemical studies. The redox couple of U(VI) to U(IV) has a reduction potential of about ~−500 mV when measured in phosphoric acid [40] . In perchloric acid, the redox couple of uranyl ion from U(VI) to U(V) was measured at −140 mV [41] . When a the uranyl ion was complexed with organic acids in acidic pH, the redox potential of the U(VI) to U(V) ranged from −270 mV to −510 mV [41] . Under environmentally relevant conditions, the redox potential of microbial terminal electron acceptors and uranyl hydroxides, chlorides, and carbonates were measured. The redox couple of U(VI)/U(IV) for these uranyl species ranged from −130 mV to −60 mV [42] , easily accessible for dissimilatory metal reducing organisms. The two electron reduction in environmental is at a slightly higher potential than to the actual proteins, which are discussed below, that the many diverse organisms use to reduce uranium. The electron transfer proteins and molecules of anaerobic organisms, such as cytochromes, quinones and blue copper proteins, typically have redox potentials in the −300 mV to −200 mV [1] . Since the proteins that reduce uranium are at a lower potential than uranyl ion complexes, the reduction reaction is thermodynamically favorable. If any of these proteins contacted the environmental uranium species in a functional configuration, electron transfer and thereby reduction would happen. The pH of the soil or groundwater and complexation ions of the uranyl ion in the environment poise the redox potential for uranium reduction at a value that is physiologically relevant for many environmental organisms. Therefore many organisms can be uranium reducers without evolving special mechanisms for uranium because they already have the electron transfer proteins at the right redox potential.
As different organisms live at different regions in the soil corresponding to the redox gradient, there is also a priority of substrates or metabolic pathways employed during uranium reduction. Also, since species of iron-reducing, sulfate-reducing, and nitrate-reducing bacteria can reduce uranium, natural communities including these bacteria can switch between active metabolic states based on availability of electron donors and acceptors while maintaining uranium-reducing conditions. While studying which members of the Geobacteracae family reduce uranium, it was observed that uranium was reduced under iron- reducing and sulfate-reducing conditions, but iron reduction happened first and then the community switched to sulfate reduction [87] . This priority order can be attributed to the redox and energy potentials of Fe(III) and sulfate, with iron being more energetically accessible. Another study under ethanol amended conditions found the same priority order of substrates, electron acceptors, nitrate, ferrous iron, sulfate and finally methane production, where 60% of the uranium reduction occurred during iron reduction phase [88] . Fe(III) amendments have also been shown to decrease the population of sulfate-reducing bacteria while stimulating uranium reduction [89] , while in other cases, the ferrous iron amendments were reported to limit uranium reduction [90] . After sulfate reduction, phases other than methanogenesis have been observed where Firmicutes dominate and carry-out biosorption rather than reduction on the remaining uranium [91] . An unusual example is the sulfate-reducing strain UFZ B 490, which can grow under nitrate-reducing conditions and still reduce uranium, but prefers to grow with sulfate [50] . Uranium reducers represent a diverse group of organisms that are found globally and at a wide span of redox potentials in the environment.
Figure 1. Mechanisms of uranium reduction. Mechanisms of uranium reduction. Reduction by: 1) Conductive pili or pili with cytochrome proteins; 2) Precipitation on outer- membrane; 3) Outer-membrane cytochrome proteins; 4) Electron flows coming from quinone pool or hydrogenases and leading to periplasmic or outer-membrane cytochrome proteins; 5) Precipitation with periplasmic cytochrome proteins; 6) Transport through porins and efflux proteins; 7) Thioredoxin involved cytoplasmic reduction path- way; 8) Uranium damaging DNA through oxidative damage.
2.1.2. Mechanisms of Uranium Reduction
There are several proposed mechanisms of uranium reduction that vary significantly by species, suggesting that there are no unique, special or evolved proteins specifically for uranium reduction, but rather a process that can happen when the conditions are right. Since, uranium is a heavy metal with redox dependent solubility, many mineral forms and many soluble chemical species exist making the landscape of reduction mechanisms complex and highly variable by organism and environmental condition (Table 3). However variable, all the mechanisms are a two electron reduction process. Studies on the mechanism of uranium reduction in Geobacter have suggested a one electron reduction to a pentavalent intermediate as a distinct pathway from the standard two electron reduction [92] . The U(V) is short lived and goes on to become U(IV) either by a second reduction step or by disproportionation of 2U(V) [92] . Density Functional Theory calculations have also indicated a U(V) intermediate originating from a Geobacter cytochrome-based uranium reduction method that also finishes with a disproportionation step [75] . Differences in intermediate states will help distinguish mechanisms in many species in the future. The variety of uranium reduction mechanisms employed that have been studied biochemically and genetically are further supported by evidence from the end point mineral forms of the reduced products. Gram-positive bacteria had different mineral endpoints than Gram-negative. Tests found a variety of carbon complexing minerals or small particulate uranium forms produced with Gram-positives [59] . These results suggest that since different genera use different mechanisms of uranium reduction, they may produce different reduced uranium minerals. However, diverse Gram-negative dissimilatory metal-reducing bacteria all produced structurally similar, but not identical, uraninite indicating that similar mechanisms may be employed by different proteins in metal-reducing bacteria [62] .
The dominant mechanism for this two electron reduction involves the donation of electrons from cytochrome c proteins to soluble U(VI). Several cytochrome and heme containing proteins have been implicated in uranium reduction. This mechanism is not altogether surprising if we consider the abiotic reactions of uranium and iron. As inorganic metals in the environment, iron in the ferrous form, alone or as part of secondary minerals, can readily reduce uranium [93] [94] . Reactions and remediation strategies relying on iron-oxide minerals, zero- valent iron or iron nanoparticles have been used to reduce and therefore immobilize uranium (Table 3) [74] [95] . Iron sulfides can also reduce uranium [96] . It follows then that in a biological system many of the proposed proteins involved in uranium reduction are multiheme cytochromes with fairly low reduction potentials.
Outer membrane and periplasmic cytochromes have been implicated strongly the uranium reduction in some organisms (Figure 1.3-1.5). Notable cytochromes are the octaheme outer-membrane cytochrome OmcZ from Geobacter, decaheme MtrC from Shewanella and tetraheme cytochrome c3 from Desulfovibrio species [49] [97] [98] . The Geobacter OmcZ has a redox potential of −200 mV, and MtrC and OmcA cytochromes from Shewanella function in range +100 to −500 mV. These cytochrome redox potentials are low enough to reduce uranium. The Geobacter family genome is packed with dozens of multiheme cytochrome proteins, and several studies have investigated which cytochromes are involved in uranium reduction. In a transcriptome analysis of Geobacter uraniireducens grown in uranium contaminated soils, an upregulation of 34 cytochrome genes was documented, several of which had been implicated in the reduction process [99] . Deletion of certain outer membrane cytochromes in Geobacter could diminish uranium reduction by as much as 60%, but was not true for all outer membrane cytochromes, as deletion of omcB or omcC did not alter the rate or extent of uranium reduction [32] . Periplasmic cytochrome MacA, a diheme c-type cytochrome peroxidase also in Geobacter was needed for any U(VI) reduction and up to 30% of the reduced uranium was found accumulated in the periplasm [97] . Recently, proteomics analysis identified one of the Geobacter cytochromes involved in uranium reduction as GscA [100] . In Shewanella, which also a heme-rich organism, outer membrane cytochrome UndA is thought to be involved in metal reductions, including uranium [101] . In Desulfovibrio species, uranium reduction pathways involve the type 1 tetraheme cytochrome c3 and evidence of precipitates of cytochrome c3 and uranium were found [47] [102] . Cytochrome proteins from Geobacter, Shewanella and Desulfovibrio species have been shown to be crucial for uranium reduction.
For the outer membrane cytochromes, the mechanism of reduction is simply the idea that a uranium molecule has access to the source of electrons and they are transferred solely based on the principles of redox chemistry (Figure 1.3). For the periplasmic soluble cytochromes, uranium must first pass through the outer membrane (Figure 1.6). Generally this movement is considered to occur through porins that allow molecules less than about 900 Da to pass independently of specific transport functions [103] . What chemical species of uranium is entering the cell and through what transporter or pore remains to be determined. The uranium atom alone has a similar size to zinc, but environmentally most uranium is bound as a soluble species, usually with carbonates, which would make oxy-ion transporters more likely. Nevertheless, when uranium randomly contacts a low-potential cytochrome in the periplasm, reduction is expected to happen spontaneously. Interestingly, as is observed with some mineral forms in nature, after the reduction, some of the cytochrome proteins are precipitated with the now insoluble U(IV) and complexes have been observed by TEM lining the inner-membranes of the periplasm (Figure 1.5) [47] [102] .
More recent work has uncovered another non-metal dependent mode of uranium reduction involving the pili proteins of Geobacter (Figure 1.1). Work from the Reguera group has shown that the pili enhance the capacity of the cells to immobilize uranium and that most uranium precipitates on the pili protecting respiratory functions operating in the periplasm and cytoplasmic membrane [104] . Without the pili, more uranium appeared to precipitate in the periplasm, likely damaging the cell (Figure 1.2) [43] . Furthermore, the pili-precipitated uranium was mononuclear with carbon-ligands; whereas, periplasmic uranium had phosphate ligands, consistent with different reduction mechanisms in the different areas of the cell [43] . The periplasmic U(VI) mineralization is likely mediated by the periplasmic facing outer-membrane cytochrome-containing membrane protein complexes [104] . The pili mechanism of reduction has been a major development in the understanding of uranium reduction mechanisms, but more broadly in bacterial extracellular electron transfer [105] . The pili have such a geometry of charge-friendly amino acid side chains that they are able to conduct electrons and deliver them to uranium which binds to the pili reversibly in negatively-charged metal binding pockets [106] . This is a very beautiful mechanism that may contribute the majority of uranium reduction in organisms with the special-architectural pili because the pili extend much further off the cell into the extracellular matrix. As with any new mechanism, it is not without its critics who maintain that cytochromes must be present for reduction to happen [107] . Supporting the conductive pili mechanism, the pili were found to enhance uranium reduction within Geobacter biofilms [108] [109] .
Intracellularly, an unusual thioredoxin containing operon has been proposed to be responsible for uranium reduction in Desulfovibrio alaskensis G20 (Figure 1.7). The Krumholz lab identified this operon by screening a transposon library for uranium reduction [110] . A strain with an interruption of the putative regulator of this operon did not show uranium reduction ability. In the wild-type strain, cadmium, a traditional thioredoxin inhibitor [111] , also shut down uranium reduction [110] . These results stimulated the interpretation that it was the thioredoxin in conjunction with a putative oxidoreductase, both encoded in the operon, that were responsible for the reduction [112] . This proposed mechanism has not been tested in vitro and the transposon mutant was not complemented with the mutated gene to show that the loss caused by the insertion was responsible for the lack of uranium reduction. A knock-out deletion of the Desulfovibrio vulgaris Hildenborough homolog of the putative thioredoxin regulator has been made that was still able to reduce uranium at approximately the same level as the parent strain (Majumder & Wall, unpublished results). There remains controversy about the transport of large quantities of uranium into the cytoplasm where thioredoxins are found. There is ample transmission electron microscopy evidence for the precipitation of uranium on cell surfaces and intraperiplasmically in many families of bacteria [25] [72] [73] [107] [111] , but fewer reports of cytoplasmic reduction sites. However, early studies of uranium uptake in Streptomyces longwoodensis did find uranium in the cytoplasmic fraction [84] . High resolution microscopic techniques like Scanning Transmission Electron Microscope-High-Angle Annular Dark-Field have also shown uranium in the cytoplasm of bacteria [64] . Since there is evidence in other bacterial species of uranium accumulation in the cytoplasm, the thioredoxin-involved uranium reduction mechanism remains feasible.
In addition to individualized mechanisms of uranium reduction, mixed or syntrophic cultures have been observed to reduce uranium [113] . Combinations of biotic and abiotic mechanisms have been shown to occur in sediment [113] . Examples of mixed biotic and abiotic systems result in different crystal forms of U(IV) precipitated. Biofilms with both reduction processes and a purely abiotic system (with ferrihydrite, sulfide and azide-inhibited Desulfovibrio) resulted in a nanocrystalline form of U(IV) that was more easily oxidized than purely biotically formed U(IV) [114] . Sulfur was also found in the crystals indicating that sulfide had been oxidized [114] . Free sulfide has a redox potential that could drive uranium reduction. On iron mineral surfaces, sulfide can compete with adsorbed U(VI), mobilizing the uranium and enabling biotic or abiotic reduction [96] . Thus sulfate-reducing bacteria producing sulfide could facilitate uranium reduction by liberating adsorbed U(VI) and reducing the metal. When a
Table 3. Complexes of uranium.
microbial community was grown on activated carbon, which increases reaction surface area, with immobilized iron nanoparticles, Baiget et al. reported 96% reduction in just 30 minutes [115] . Mixed cultures or communities were also screened for uranium reduction, usually testing natural communities from uranium contaminated sites. In these situations, the individual organisms contributing to the uranium reduction were not identified [10] [29] [51] [116] . From all of these examples, uranium reduction was observed abiotically, extracellularly, periplasmically, cytoplasmically and in mixed biotic-abiotic systems and the mechanisms examined reveal a diverse array of uranium reduction reactions from a diverse set of bacteria (Table 2 & Table 3). The diversity in uranium reducing organisms and reduction mechanisms suggests that uranium reduction is a more universal process and driven by chemical conditions rather than biological benefit.
2.1.3. Factors Influencing Reduction Reaction Rates & Electron Flow Pathways
Multiple factors affect reduction rates in microbes meaning that certain uranium complexes, energy sources for the organisms and electron flow pathways are more efficient at reducing uranium. The bioavailability of a metal is a major determinant of what bio-transformation happens and at what rate. Uranium in the hexavalent oxidation state is soluble and when reduced to the tetravalent oxidation state, it is insoluble. Uranium is more soluble at low pH and with increasing concentrations of anions like carbonates that form soluble complexes with the metal [117] . Additionally, calcium-carbonate-uranium(VI) complexes have an even greater decrease in bio-availability, but an increased solubility suggesting that calcium is an inhibitor of uranium bio-transformations (Table 3) [48] [118] . The ending mineral form is also indicative of which mechanism or transformation reaction happened and dictates what happens to the metal, for example, does it stay immobilized? Is the metal accessible for additional bio- or chemical transformations? Insoluble U(IV) can precipitate as the simple mineral uraninite, UO2, but more often forms minerals with calcium and phosphates such as autunite [119] . A more labile precipitate has also been identified known as monomeric U(IV) [120] [121] .
In addition to solubility, complexation or speciation of the uranium metal impacts reduction rates. Different species of bacteria are able to reduce different hexavalent U(VI) organic acid complexes and with varied reduction rates. D. desulfuricans reduced monodentate aliphatic complexes, like acetate, more rapidly while Shewanella alga reduced multidentate complexes, like citrate, more rapidly [122] . Likewise, uranium in soils is often adsorbed or associated with surfaces on the soil which have been shown to limit bioavailability and decrease reduction rates [123] . Geobacter species were still able to reduce uranium under phosphate-limiting conditions, with the increased bio-availability of U(VI) possibly accounting for decreased cell growth [124] . Still others report non-uraninite minerals as the major form of uranium trapping in an environmental setting because phosphoryl ligands can inhibit U(VI) bioreduction. Thus there is a need for considering uranium speciation and what chemical species are environmentally relevant for further testing and remediation attempts [23] . Uranium (VI) complexed with hydrogen and phosphate could be more easily reduced by several metal reducing species when adsorbed to non-reducing Bacillus subtilus [61] . In other studies, Shewanella putrefaciens easily reduced uranyl-carbonate complexes, but reduced virtually no uranium when presented with calcium-uranyl- carbonate complexes [125] . Increasing bicarbonate slowed uranium reduction kinetics in Shewanella oneidensis by changing the chemical speciation [126] . Additional studies show the limitation of uranium reduction with increased calcium and iron species, like ferrihydrite and iron (III) oxides, concentrations which affect the speciation of soluble uranium complexes [18] [127] [128] . Like solubility and speciation, the reduction potential of U(VI) is dependent on pH, metal ion-complexation and concentration. An increase in organic acid complexation or pH decreases the redox potential [41] . Therefore, uranium reduction only happens when the pH, complexation and concentration are just right to tune the redox potentials and form bioavailable uranium.
While the influence of the electron donor on reduction rate is well-studied, the electron source can also control microbial community members. For someClostridial species, fermentative growth was reported to be important for uranium reduction and, in the same study, an increase was noted with more hydrogen [129] . This signaled the importance of the hydrogenases in electron and energy flows that terminate with uranium reduction (Figure 1.4). Neither nitrate nor nitrogen gas affected reduction rates, but pH did impact rates with pH 4 being optimal in Clostridia sp. [35] [129] . Similarly for Shewanella species, hydrogen as the electron donor yielded faster reduction rates, 110 μmoles/(h・108 cells/mL), than lactate [130] [131] ; whereas, non-growing cells had a rate of 2.37 µmoles/(h・mg biomass) [52] . Further evidence for the importance of hydrogenases comes from South Africa. The ability of a microbial community to reduce uranium was inhibited by rotenone, a NADH-dependent hydrogenase inhibitor, suggesting that electrons going to uranium were coming through hydrogenase [111] . Hydrogenases were also found to be part of the uranium reduction pathway from Desulfovibrio species going through the periplasmic cytochrome c3 [47] . Rates with organic acids as the electron donor were half that of those with hydrogen [47] . The addition of electron carrier molecules, so called electron shuttles, such as quinones like anthraquinone-2,6-disulfonate, have also increased uranium reduction rates by increasing electron flow [90] [132] [133] . In Shewanella species FMN (Flavin mononucleotide) was found to be an important electron transfer molecule in the uranium reduction pathway [134] . On a community scale, during electron donor limited conditions in environmental setting lesser known organisms such as Pelosinus outcompeted the traditional metal reducers and maintained metal reduction activity [135] . As with bioavailabilty of the metal, electron and carbon sources also impacted microbial ability to reduce uranium and the reduction rates.
The organisms, the mechanisms and the rates of uranium reduction have been extensively studied but are not fully characterized. As more about uranium reduction is uncovered, other bio-transformations of uranium are being discovered.
2.2. Uranium Oxidation
Uranium oxidation involves taking U(IV) and oxidizing it to U(VI), the exact opposite of the reduction process (Figure 2.6). Typically, this happens spontaneously when oxygen invades the anaerobic environment, but there is new evidence for bacteria that oxidize uranium. To date, only two organisms have been characterized as uranium oxidizers, Geobacter metallireducens and Thiobacillusdenitrificans (Table 2) [86] . G. metallireducens is a well-known uranium reducer so it was surprising that it has also been shown to oxidize uranium; it is possible that the non-reducing cytochromes may play a role in oxidation. In the nitrate-dependent uranium oxidation by β-Proteobacteria sulfur oxidizer and nitrate reducer T. denitrificans two diheme cytochrome c’s accounted for at least 50% of the oxidation capacity [86] . With the addition of nitrate or active denitrification, no uranium reduction was detected suggesting nitrate amendment may stimulate uranium bio-oxidation [136] . Another study [137] , used 16S rRNA profiling to look for changes in the bacterial community during uranium reduction and the reoxidation that is observed at some sites over time. Brodie et al. found that metal-reducing members such as Geothrix fermentas and Geobacteraceae grew during the uranium reduction phase and persisted during reoxidation suggesting that these microbes could be responsible for the reoxidation [137] . The individual and community studies taken together present a stronger case that microbes may be causing at least some of the reoxidation of uranium in the environment, particularly any reoxidation observed under anaerobic conditions. It is possible that some communities are forming a uranium cycle as an electron acceptor. However as is the case with reduction, oxidation could be primarily a chemistry-driven process.
2.3. Uranium Respiration
Anaerobic organisms grow by respiring a variety of electron donors and acceptors. Many microbes respire organic acids, oxyanions such as sulfate and nitrate, metals and other electron acceptors available to them in their environment, if they can extract an electron and then find an acceptor to give it too, they will do it (Figure 2.5). These anaerobic metabolisms drive important geochemical cycles on the Earth such as Sulfur, Nitrogen and Carbon Cycles. A big question in the study of uranium bio-transformations has always been whether these microbes are respiring uranium, particularly in the cases of uranium reduction. Do organisms gain energy from that process and grow, or was uranium just the closest convenient electron dump for the excess of electrons often built up in anaerobic microorganisms?
Experimentally, proving uranium respiration requires rigorous evidence and is difficult. At the Rifle site, isolates with sequence similarity to Burkholderia fungorum were deduced to respire uranium using acetate as the electron donor [66] . Acidithiobacillus ferrooxidans respiration rates were altered by changing uranium concentrations, but it is unclear if this bacterium was respiring uranium [56] . Anaeromyxobacter dehalogenans is predicted to respire uranium and a correlation was found between cell number and the uranium found in wells at the Oak Ridge [33] . S. putrefaciens and its mutants were found to respire a number of electron acceptors with a few strains capable of anaerobic growth on uranium [54] . Some Geobacter strains have been found to respire uranium as well [138] . Uranium respiration has not been shown yet by the standard microbiological techniques where cells are grown with sub-inhibitory concentrations of uranium as the only potential electron acceptor with incremental additions and growth. It would certainly not be a ‘normal’ metabolism. Under duress microbes have been found to grow in amazing circumstances and studies are pointing to uranium respiration being possible under the right conditions.
2.4. Detoxification/Resistance
There is an on-going debate in the field regarding the purpose of uranium bio-transformations for the microbe. Are these metabolic changes part of a detoxification and resistance effort against the toxic, heavy metal, or radionuclide or are these processes that occur because of environmental circumstance? In many instances in the literature, uranium reduction is liberally referred to as detoxification [50] [67] [68] [79] [80] [139] , but this nomenclature may not always be accurate from the perspective of the microbe. First of all, uranium is toxic because it causes oxidative damage like other heavy metals (Figure 1.8) [15] . The radioactivity from natural abundance and depleted sources of uranium is barely above background, causing few health risks.
For the microbes that reduce uranium, in some cases, bio-transformations protect cells by removing the toxic metal or sequestering it in the extracellular matrix [68] [82] . However, some of the processes cause solid uranium buildup on cell surfaces, in the periplasm and possibly even the cytoplasm [47] [102] [110] . This build up should eventually obstruct normal cellular function and is noted anecdotally, but lacks concrete evidence. Park and Jiao reported that C. crescentus exhibits growth arrest and cessation of DNA replication when first exposed to uranium [67] . In the same study, they describe a novel mechanism in which C. crescentus raises the pH of the solution making U(VI) less soluble and consequently less toxic [67] , which differs from other detoxification strategies which involve reduction and precipitation. Yung et al. studying the same species found in uranium-exposed cells an upregulation of an ABC-type transporter interpreted as possibly providing efflux for detoxification [69] . The same lab followed up with a Tn-seq (transposon sequencing) study and found evidence for the involvement of additional transporters and stress-associated transcription factors that facilitated survival with uranium [70] . In the case of Geobacter, proteomics of Geobacter cells grown in the presence of uranium revealed an increase in proteins for efflux pumps and proteins that handle oxidative damage, suggesting that as for bio-reduction, there is no mechanism specific to uranium enzymatic processes for detoxification [140] . Global transcriptional profiling of S. oneidensis MR-1 during uranium reduction revealed several classes of genes that were upregulated. Genes known for accessing alternative electron acceptors such as manganese, soluble iron, and fumarate were upregulated along with genes for dealing with stress and damage to membranes [53] . The upregulation of alternative respiratory genes is supported with other evidence that certain energy flows are more favorable for uranium reduction, but these alternative pathways are not unique to uranium stress. However, the upregulation of membrane damage response proteins could be seen as a response of the cell to remove the uranium. The gene changes in response to stress speaks to the oxidative damage caused by the heavy metal toxicity associated with U(VI). However, in responses where U(IV) precipitates on the outer membrane or inside the periplasm, sometimes associated with vital electron shuttle cytochromes, there may also be toxic effects. The precipitated heavy metal is in permanent direct contact with the cell. In the case where the uranium is precipitated away from the cell, the physical removal and immobilization can be argued to be less toxic. So based on gene transcription responses and location of precipitated uranium, the reduction process could be considered a detoxification response if the genes for reduction are upregulated and the uranium is precipitated away from the cell. Experimental verification remains to confirm this hypothesis about detoxification and reduction.
2.5. Chemical and Biological Drivers of Uranium Reduction
Having explored the reactions and mechanisms of uranium reduction, we ask if chemistry or biology is the primary driver of this process. In other examples of microbial metal bio-transformations (reduction, resistance) there are specific and conserved sets of genes that express proteins to chemically transform certain metals. Those proteins include CzcA, copper antiporter, Acr3, arsenic dissimilatory reductase, and HgcAB, mercury methylation [141] [142] [143] . No designated or uranium-specific conserved proteins have been found to date. So far, all implicated proteins have other primary functions and appear to do uranium reduction when the opportunity presents itself. Several different mechanisms of uranium reduction have been observed (Figure 1) with different starting uranium complexes and different mineral endpoints (Table 3), suggesting a lack of similarity across organisms or mechanisms. Uranium-reducing organisms have been found all over the planet and in multiple types of environments. Uranium reducers are from many families of organisms and even include aerobes and non-dissimilatory metal reducers (Table 2). Some organisms may respire uranium and have slight energy gains, but there is little or no biological advantage to the microbes with the uranium reduction mechanisms as described since most were measured during stationary phase. The purpose of uranium reduction does not appear to be for detoxification either. Therefore, we argue that based on reduction potential, the global nature and occurrence of uranium reduction in many types of Earth environments, the variety of mechanisms, non-conserved mineral endpoints and the lack of specifically conserved enzymes, that the process of uranium reduction is predominantly driven by chemistry, meaning the spontaneous movement of electrons drives the reaction rather than an enzymatically catalyzed process. It seems to be a more universal process, that any microbe in the right environment would be capable of reducing uranium. As other uranium bio-transformations are explored, it is possible that biology rather than chemistry may be found to be the driver.
3. Binding, Sensing and Synthesizing Bio-Transformations of Uranium
3.1. Sorption
Bio-sorption of uranium is the process where the uranium is immobilized on the outer membrane or extra polymeric substance of the microbe (Figure 2.9). This can follow a redox event, but can also be redox independent. It can likewise sometimes rely on charge, but can also be due to electrostatics since uranium is “sticky.” Sorption is one of the more commonly referenced bio-transformations in the literature because it is a possible remediation strategy. Some cells can adsorb uranium up to nearly half their cell weight [84] [85] , but this appears to be a passive, non-enzymatically driven process.
Examples of microbes carrying out biosorption include the fungus Talaromyces emersonii. At pH 5, the fungus had a biosorption capacity of 280 mg U per gram of dry cell weight [85] . With maximal biosorption of 0.44 g U per gram of dry cell weight at pH 4.6, the Actinomycetes bacterium Streptomyces longwoodensis efficiently removes uranium from liquid samples [84] . The amount was dependent on several factors including cell phosphorous content, pH, uranium concentration, and cell-cycle stage [84] . Bacillus isolates from Saxony were able to take up uranium efficiently in the 10 - 200 mM range and the metal was found adsorbed to the S-layer proteins and cell surface [144] . In a surprising
Figure 2. Mechanisms of bio-transformation of uranium. Mechanisms of bio-transformations of uranium. 1) Mineralization―the formation of uranium-containing minerals with carbonates, calcium, enzymatically cleaved phosphates or other molecules. 2) Nanoparticles―synthesis of uranium nanoparticles may be uraninite or other mineral forms. 3) Precipitation―deposition of insoluble uranium alone or in mineral form. 4) Accumulation―concentration of uranium inside at least one membrane. 5) Respiration―growing or gaining of energy from uranium reduction. 6) Reduction/Oxidation―uranium gaining or losing two electrons 7) Marker―isotopic fractionation of uranium by microbe. 8) Sensing―detection of uranium by binding of a molecule that releases a fluorescent molecule. 9) Sorption―uranium binding to outer membrane, EPS, S-layer, filaments, pili or nanowires.
finding, Thorgensen and co-workers identified an S-layer protein complex in Pelosinus sp. Strain UFO1 that binds U(VI), but does not reduce it [77] . Diverse organisms such as At. ferrooxidans and Acidovorax facilis are also reported to adsorb uranium [56] [57] . Even non-uranium-reducers or organisms not found in uranium contaminated environments, such as Saccharomyces cerevisiae and Staphylococcus aureus LZ-01, can adsorb uranium efficiently [79] [83] . The chemical species of the adsorbed uranium varies and reflects the state of uranium in the environment. Uranium is often tightly bound, complexing to phosphoryl, carbonate or occasionally nitryl ligands and side chains. Therefore biosorption can be an attractive method for bio-remediation.
3.2. Accumulation
Accumulation can sometimes be referred to interchangeably with bio-sorption. However, sorption and accumulation are generally differentiated by the location of the bio-transformed uranium relative to the cell. Accumulated uranium is located within the cell, either in the periplasm or cytoplasm, but must have traversed at least one membrane (Figure 2.4). Like biosorption, Cellulomonas [72] and fungi [16] have been shown to accumulate impressive amounts of uranium. As discussed in the reduction section, the mechanism of uranium entry into the cell is unknown. It is also not well-characterized how the accumulation of uranium impacts cell growth and survivability, but the suggestion is that it is detrimental or neutral at best, but does not apparently benefit the cell.
3.3. Mineralization
Bio-mineralization is also more generally defined as the formation of U(IV) species, which is insoluble and called the mineral uraninite, UO2. However, this is the end product of bio-reduction, so the terms are sometimes used interchangeably. Bio-mineralization can also be more specifically defined as an active enzymatically driven process where the uranium is transformed to insoluble non- oxide mineral, usually metal phosphates (Table 3) [139] . The mechanism of bio-mineralization has been well-characterized in the γ-Proteobacteria Serratia sp. [145] . Uranium is trapped on the surface of the cell or in the extracellular matrix on material like EPS, extracellular polymeric substances (Figure 2.1). Meanwhile, the enzyme phosphatase, that can be found on the outer membrane and can also be excreted into the extracellular matrix, creates inorganic phosphate by hydrolyzing the phosphate from soluble phosphate organic compounds or minerals. The surface-complexed uranium, phosphate and often calcium react forming immobilized minerals [81] [82] [145] . In Serratia this is seen under fermentative growth and different minerals may be produced under phosphate limited conditions [82] .
In addition to Serratia, the C. crescentus bio-mineralization mechanism is also well studied and the specific periplasmic phosphatase releasing phosphate, PhoY, has been determined [68] . Citrobacter sp. also bio-mineralizes uranium on the cell surface forming phosphate minerals with phosphate, while ammonium addition to the mineral produces an even more insoluble form [73] . Arthrobacter sp. and Microbacterium oxydans from mine tailing sites were also found to produce uranium phosphate minerals, confirmed by High Resolution Transmission Electron Microscopy [64] .
Uranium can be mineralized into a variety of uranyl-phosphates by active microbes. The term bio-precipitation (Figure 2.3) is also used when the specific process is unknown, but solid uranium particles are observed such as in a batch community system or environmental field site [82] [94] [146] [147] . A well- known bio-precipitation is mediated by Citrobacter sp., which forms slightly yellow uranyl phosphates [73] .
3.4. Bio-Markers of Uranium Bio-Transformations
Bio-markers are small molecules, genes, proteins or even isotope fractionation ratios that consistently correspond with a biologically-driven process. Isotope fractionation is the process where microbes preferentially consume a specific isotope of an element creating an enrichment of the preferred isotope in the end product that differs from the natural abundances of the isotopes (Figure 2.7). The enrichment is usually a constant ratio. If isotope fractionation values are known for certain biologically-driven processes and that fractionation value is found in samples, it is strong evidence that the process occurred biologically even if the bacteria responsible are no longer present, such as in deep cores from early Earth eras.
Bio-makers have been used to detect uranium biotransformation that took place millions of years in the past. Uranium reduction does have a known, although difficult to measure isotope fractionation value that actually enriches the heavy isotope [60] [148] [149] . Rademacher and coauthors compared the isotope fractionation for uranium reduction from an abiotic process with zero valent iron particles and bioreduction by Geobacter or Anaeromyxobacter. They found that the biotic process produced a very similar negative epsilon (enrichment factor) value for both bacteria, while the abiotic iron process resulted in the same ratio as natural abundance [60] . Dang et al. also compared biotic and abiotic pathways and came to the same conclusion, but with the addition that certain minerals such as mixtures of iron and manganese have an isotope preference for the lighter isotope, U-235 [148] . The Bernier-Latmani group has also measured a uranium signature with preference for the heavy isotope and has used this signature to detect microbial activity in rocks from multiple ages of Earth history including Archean and Cretaceous [149] . A proposed biomarker for Geobacter is the amount of the genus specific GltA protein [150] . Wilkins et al. showed that there was a strong correlation between copy numbers of the protein versus uranium reduction activity during stimulated bioremediation [150] . With modern technology, ancient and current uranium bio-transformations are detectable.
3.5. Bio-Sensors
Knowing the nature of the chemical interaction between microbes and uranium has enabled the creation of bio-sensors for uranium in the environment that have taken a variety of forms. Bacterial community composition has even been used as a geo-sensor to predict the location of uranium contamination [151] . Antibodies to certain uranium chemical species have also been developed as an environmental biosensor [152] .
UO2-selective DNA-zymes, which are engineered pieces of DNA designed to specifically bind the uranyl ion and then cleave a fluorescent molecule containing DNA arm, releasing the molecule which emits fluorescence (Figure 2.8) [153] [154] . Quenching the fluorescence with a 2D layer of molybdenum disulfide nanosheets allows the decrease in fluorescence to be a readout for U(VI) concentration [154] .
C. crescentus has been used as a whole cell uranium biosensor by engineering the organism to produce fluorescence when excited with UV light and when its environment contains uranium. As a fluorescent biosensor, this bacterium has a detection limit in the nanomolar range [37] . Bio-sensing presents an in situ method for uranium detection in the environment.
3.6. Biofilm/Biobarrier
Bacterial biofilms are sessile tight assemblages of a microbial population that create micro-environments or niches that may be different than the surrounding environment. Biofilms have shown enhanced uranium sequestration and immobilization in single organism and multi-organism biofilms. Biofilms of strains of iron-reducing Shewanella species were found to immobilize uranium efficiently, and Shewanella extracellular polymeric substances played a very large role in that immobilization with the assistance of reduction by the outer-membrane cytochromes [55] . However, reduction capability was dependent on biofilm architecture and the reactor used to measure that structure [155] . Examination of Geobacter biofilms saw increased uranium resistance up to 5 mM uranium and better immobilization compared with planktonically grown cultures; the conductive pili and cytochromes were major contributors to the biofilms enhanced resistance [108] . Desulfovibrio biofilms routinely formed nanocrystalline U(IV) over other crystal types formed by planktonic cultures [114] . Looking at community-scale biofilms, rhizosphere communities growing on iron-plaques in the DOE Savanah River Site were also found to have enhanced uranium reduction and immobilization [156] . Stemming from the increased immobilization with biofilms, some remediation strategies include biofilms as a form of bio-barrier to trap the uranium or are working to develop the biofilms to deploy in permeable reactive barriers [65] [108] [157] . New laser-based fluorescence techniques have been employed to determine the chemical species of uranium in the biofilm environment [158] . Biofilms characteristically have increased interactions between the microbes which results in altered or enhanced interactions with uranium.
3.7. Nano-Particle Synthesis, Nano-Wires
In the bacterial produced nano-realm, nano wires, fibers and particles have been described with reduced uranium (Figure 2.2). Nano-wires/fibers of meta-schoe- pite mineral polycrystalline chains were produced by S. oneidensis MR-1 during uranium reduction (Table 3). Another reduction reaction could reduce these nanowires to nanoparticles [159] [160] . Cytochromes were found to be necessary for nanoparticle U(IV) formation by Shewanella [98] . Structural characterization of Shewanella biogenic uraninite nanoparticles suggested that the synthesis pathway was thermodynamically driven [161] . The long conductive pili of Geobacter with and without cytochromes have also been called nanowires because of their ability to transmit electrons [13] . The bio-produced nanomaterials have benefit to the cells and could be used by humans for remediation or other electronic applications.
4. Conclusion and Outlook
4.1. Contributions of Inorganic Chemistry to the Study of Uranium Bio-Transformations
While perhaps not widely recognized, standard inorganic chemistry techniques and their advancement have been integral to the characterization and determination of the uranium bio-transformations on a molecular level. The following techniques have been used extensively in the references of this review paper: Mossbauer spectroscopy, X-ray Absorption Spectroscopy (XAS), Extended X-ray Absorption Fine Structure (EXAFS), X-ray Absorption near Edge Structure (XANES), X-ray Powder Diffraction (XRD), X-ray Photoelectron Spectroscopy (XPS), Transmission Electron Microscopy (TEM), Atomic Force Microscopy (AFM), Density Functional Theory calculations (DFT) and laser induced fluorescence spectroscopy. These techniques determine the chemical species of the uranium in the solid mineral, soluble complexes, complexed and ligated to surfaces, mechanism intermediates, oxidation state and more. They have revealed how the important chemical speciation is to the biological process. As with any chemical reaction, certain ligands or chemical forms are highly favorable for reaction and others are inhibitory or energetically inaccessible. Knowledge of these chemical details from inorganic studies have transformed how microbiologists think about uranium bio-transformations and the mechanisms by which they happen.
4.2. Chemistry or Biology?
Even though uranium is a highly toxic metal, microbes have developed the ability to transform uranium in a number of ways. Microbes reduce, oxidize, possibly respire, mineralize, precipitate, and accumulate uranium. Humans take advantage of the reactions to stimulate remediation of contaminated sites, to sense uranium in the environment, to determine the biological activity in the early earth, or to synthesize nanoparticles and wires. The variety of interactions with uranium leaves many areas of research still open: characterization of more reduction mechanisms, identifying more organisms capable of these processes, understanding uranium-reducing community dynamics on a molecular scale, determining if microbes respire uranium, determining the biological benefit to the microbes that do uranium bio-transformations, and determining if and how microbes survive with accumulated and precipitated uranium in and on cells. Underlying all of it is the classical question in Earth’s processes―chemistry or biology or an intriguing combination?
Acknowledgements
This material by ENIGMA―Ecosystems and Networks Integrated with Genes and Molecular Assemblies (http://enigma.lbl.gov), a Scientific Focus Area Program at Lawrence Berkeley National Laboratory is based upon the work supported by the U.S. Department of Energy, Office of Science, Office of Biological & Environmental Research under contract number DE-AC02-05CH11231.
Cite this paper
Majumder, E.L.-W. and Wall, J.D. (2017) Uranium Bio-Transfor- mations: Chemical or Biological Processes? Open Journal of Inorganic Chemistry, 7, 28-60. https://doi.org/10.4236/ojic.2017.72003
References
- 1. Bertini, I., Gray, H.B., Stiefel, E.I. and Selverstone Valentine, J. (2007) Biological Inorganic Chemistry: Structure and Reactivity. University Science Books, Sausalito, California.
- 2. Kendall, B., Brennecka, G.A., Weyer, S. and Anbar, A.D. (2013) Uranium Isotope Fractionation Suggests Oxidative Uranium Mobilization at 2.50 Ga. Chemical Geology, 362, 105-114.
https://doi.org/10.1016/j.chemgeo.2013.08.010 - 3. Newsome, L., Morris, K. and Lloyd, J.R. (2014) The Biogeochemistry and Bioremediation of Uranium and Other Priority Radionuclides. Chemical Geology, 363, 164-184.
https://doi.org/10.1016/j.chemgeo.2013.10.034 - 4. Nolan, J.P. (2016) Mobilization of Naturally Occurring Uranium in Sediment into Groundwater. Ph.D. Thesis, University of Michigan, Ann Arbor.
- 5. Panda, S., Mishra, S. and Akcil, A. (2016) Bioremediation of Acidic Mine Effluents and the Role of Sulfidogenic Biosystems: A Mini-Review. Euro-Mediterranean Journal for Environmental Integration, 1, 8.
https://doi.org/10.1007/s41207-016-0008-3 - 6. Zachara, J.M., et al. (2013) Persistence of Uranium Groundwater Plumes: Contrasting Mechanisms at Two DOE Sites in the Groundwater-River Interaction Zone. Journal of Contaminant Hydrology, 147, 45-72.
https://doi.org/10.1016/j.jconhyd.2013.02.001 - 7. Gehle, K. (2013) Uranium Toxicity.
https://www.atsdr.cdc.gov/csem/csem.asp?csem=16&po=0 - 8. Crawford, S.E., Lofts, S. and Liber, K. (2017) The Role of Sediment Properties and Solution pH in the Adsorption of Uranium (VI) to Freshwater Sediments. Environmental Pollution, 220, 873-881.
https://doi.org/10.1016/j.envpol.2016.10.071 - 9. North, N.N., et al. (2004) Change in Bacterial Community Structure during in Situ Biostimulation of Subsurface Sediment Co-Contaminated with Uranium and Nitrate. Applied and Environmental Microbiology, 70, 4911-4920.
https://doi.org/10.1128/AEM.70.8.4911-4920.2004 - 10. Cho, K., et al. (2012) Linking Bacterial Diversity and Geochemistry of Uranium-Contaminated Groundwater. Environmental Technology, 33, 1629-1640.
https://doi.org/10.1080/09593330.2011.641036 - 11. Xu, M., et al. (2010) Responses of Microbial Community Functional Structures to Pilot-Scale Uranium in Situ Bioremediation. The ISME Journal, 4, 1060-1070.
https://doi.org/10.1038/ismej.2010.31 - 12. Wall, J.D. and Krumholz, L.R. (2006) Uranium Reduction. Annual Review of Microbiology, 60, 149-166.
https://doi.org/10.1146/annurev.micro.59.030804.121357 - 13. Lovley, D.R., et al. (2011) Geobacter: The Microbe Electric’s Physiology, Ecology, and Practical Applications. Advances in Microbial Physiology, 59, 1-100.
- 14. Zhao, C., et al. (2016) Characterization of Uranium Bioaccumulation on a Fungal Isolate Geotrichum sp. dwc-1 As Investigated by FTIR, TEM and XPS. Journal of Radioanalytical and Nuclear Chemistry, 310, 165-175.
https://doi.org/10.1007/s10967-016-4797-2 - 15. Volesky, B. and Holan, Z.R. (1995) Biosorption of Heavy Metals. Biotechnology Progress, 11, 235-250.
https://doi.org/10.1021/bp00033a001 - 16. Cumberland, S.A., Douglas, G., Grice, K. and Moreau, J.W. (2016) Uranium Mobility in Organic Matter-Rich Sediments: A Review of Geological and Geochemical Processes. Earth-Science Reviews, 159, 160-185.
- 17. Lovley, D.R., Phillips, E.J.P., Gorby, Y.A. and Landa, E.R. (1991) Microbial Reduction of Uranium. Nature, 350, 413-416.
https://doi.org/10.1038/350413a0 - 18. Stewart, B.D., Girardot, C., Spycher, N., Sani, R.K. and Peyton, B.M. (2013) Influence of Chelating Agents on Biogenic Uraninite Reoxidation by Fe(III) (Hydr)oxides. Environmental Science & Technology, 47, 364-371.
https://doi.org/10.1021/es303022p - 19. Wu, W.M., et al. (2007) In Situ Bioreduction of Uranium (VI) to Submicromolar Levels and Reoxidation by Dissolved Oxygen. Environmental Science & Technology, 41, 5716-5723.
https://doi.org/10.1021/es062657b - 20. Liu, C., et al. (2005) Influence of Sediment Bioreduction and Reoxidation on Uranium Sorption. Environmental Science & Technology, 39, 4125-4133.
https://doi.org/10.1021/es048501y - 21. Komlos, J., Peacock, A., Kukkadapu, R.K. and Jaffé, P.R. (2008) Long-Term Dynamics of Uranium Reduction/Reoxidation under Low Sulfate Conditions. Geochimica et Cosmochimica Acta, 72, 3603-3615.
- 22. Bonnetti, C., et al. (2017) Coupled Uranium Mineralisation and Bacterial Sulphate Reduction for the Genesis of the Baxingtu Sandstone-Hosted U Deposit, SW Songliao Basin, NE China. Ore Geology Reviews, 82, 108-129.
- 23. Morin, G., et al. (2016) Mononuclear U(IV) Complexes and Ningyoite as Major Uranium Species in Lake Sediments. Geochemical Perspectives Letters, 2, 95-105.
https://doi.org/10.7185/geochemlet.1610 - 24. van Berk, W. and Fu, Y. (2017) Redox Roll-Front Mobilization of Geogenic Uranium by Nitrate Input into Aquifers: Risks for Groundwater Resources. Environmental Science & Technology, 51, 337-345.
https://doi.org/10.1021/acs.est.6b01569 - 25. Maleke, M., et al. (2015) Optimization of a Bioremediation System of Soluble Uranium Based on the Biostimulation of an Indigenous Bacterial Community. Environmental Science and Pollution Research, 22, 8442-8450.
https://doi.org/10.1007/s11356-014-3980-7 - 26. Newsome, L., Morris, K., Trivedi, D., Atherton, N. and Lloyd, J.R. (2014) Microbial Reduction of Uranium (VI) in Sediments of Different Lithologies Collected from Sellafield. Applied Geochemistry, 51, 55-64.
- 27. Haferburg, G. and Kothe, E. (2007) Microbes and Metals: Interactions in the Environment. Journal of Basic Microbiology, 47, 453-467.
https://doi.org/10.1002/jobm.200700275 - 28. Lovley, D.R. (2000) Environmental Microbe-Metal Interactions. ASM Press, Washington, DC.
https://doi.org/10.1128/9781555818098 - 29. Mondani, L., et al. (2011) Influence of Uranium on Bacterial Communities: A Comparison of Natural Uranium-Rich Soils with Controls. PLoS One, 6, e25771.
https://doi.org/10.1371/journal.pone.0025771 - 30. Yan, X., Luo, X. and Zhao, M. (2016) Metagenomic Analysis of Microbial Community in Uranium-Contaminated Soil. Applied Microbiology and Biotechnology, 100, 299-310.
https://doi.org/10.1007/s00253-015-7003-5 - 31. Lovley, D.R. and Phillips, E.J. (1992) Reduction of Uranium by Desulfovibrio desulfuricans. Applied and Environmental Microbiology, 58, 850-856.
- 32. Sanford, R.A., et al. (2007) Hexavalent Uranium Supports Growth of Anaeromyxobacter dehalogenans and Geobacter spp. with Lower than Predicted Biomass Yields. Environmental Microbiology, 9, 2885-2893.
https://doi.org/10.1111/j.1462-2920.2007.01405.x - 33. Thomas, S.H., et al. (2010) Unique Ecophysiology among U(VI)-Reducing Bacteria as Revealed by Evaluation of Oxygen Metabolism in Anaeromyxobacter dehalogenans Strain 2CP-C. Applied and Environmental Microbiology, 76, 176-183.
https://doi.org/10.1128/AEM.01854-09 - 34. Fletcher, K.E., et al. (2010) U(VI) Reduction to Mononuclear U(IV) by Desulfitobacterium Species. Environmental Science & Technology, 44, 4705-4709.
https://doi.org/10.1021/es903636c - 35. Gao, W. and Francis, A.J. (2008) Reduction of Uranium(VI) to Uranium(IV) by Clostridia. Applied and Environmental Microbiology, 74, 4580-4584.
https://doi.org/10.1128/AEM.00239-08 - 36. Li, X., et al. (2016) Microbial Reduction of Uranium (VI) by Bacillus sp. dwc-2: A Macroscopic and Spectroscopic Study. Journal of Environmental Sciences.
- 37. Hillson, N.J., Hu, P., Andersen, G.L. and Shapiro, L. (2007) Caulobacter crescentus as a Whole-Cell Uranium Biosensor. Applied and Environmental Microbiology, 73, 7615-7621.
https://doi.org/10.1128/AEM.01566-07 - 38. Hu, N., et al. (2016) Bioreduction of U(VI) and Stability of Immobilized Uranium under Suboxic Conditions. Journal of Environmental Radioactivity, 154, 60-67.
- 39. Pepper, I.L., Gerba, C.P. and Gentry, T.J. (2015) Environmental Microbiology. Academic Press, San Diego, CA.
- 40. Baes Jr., C.F. (1953) The Reduction of Uranium (VI) by Ferrous Iron in Phosphoric Acid Aolution: The Formal Electrode Potential of the U(IV)/(VI) Couple Tech. Inf. Service Extension, Oak Ridge, Term.
- 41. Suzuki, Y., et al. (2007) Electrochemical Studies on Uranium in the Presence of Organic Acids. Journal of Nuclear Science and Technology, 44, 1227-1232.
https://doi.org/10.1080/18811248.2007.9711366 - 42. Rinklebe, J., Knox, A.S. and Paller, M. (2016) Trace Elements in Waterlogged Soils and Sediments. CRC Press, Boca Raton.
- 43. Reguera, G. (2012) Electron Transfer at the Cell-Uranium Interface in Geobacter spp. Biochemical Society Transactions, 40, 1227-1232.
https://doi.org/10.1042/BST20120162 - 44. McGuinness, L.R., Wilkins, M.J., Williams, K.H., Long, P.E. and Kerkhof, L.J. (2015) Identification of Bacteria Synthesizing Ribosomal RNA in Response to Uranium Addition During Biostimulation at the Rifle, CO Integrated Field Research Site. PLoS One, 10, e0137270.
https://doi.org/10.1371/journal.pone.0137270 - 45. Wilkins, M.J., et al. (2009) Proteogenomic Monitoring of Geobacter Physiology during Stimulated Uranium Bioremediation. Applied and Environmental Microbiology, 75, 6591-6599.
https://doi.org/10.1128/AEM.01064-09 - 46. Yun, J., Ueki, T., Miletto, M. and Lovley, D.R. (2011) Monitoring the Metabolic Status of Geobacter Species in Contaminated Groundwater by Quantifying Key Metabolic Proteins with Geobacter-Specific Antibodies. Applied and Environmental Microbiology, 77, 4597-4602.
https://doi.org/10.1128/AEM.00114-11 - 47. Payne, R.B., Gentry, D.M., Rapp-Giles, B.J., Casalot, L. and Wall, J.D. (2002) Uranium Reduction by Desulfovibrio desulfuricans Strain G20 and a Cytochrome c3 Mutant. Applied and Environmental Microbiology, 68, 3129-3132.
https://doi.org/10.1128/AEM.68.6.3129-3132.2002 - 48. Brooks, S.C., et al. (2003) Inhibition of Bacterial U(VI) Reduction by Calcium. Environmental Science & Technology, 37, 1850-1858.
https://doi.org/10.1021/es0210042 - 49. Lovley, D.R., Widman, P.K., Woodward, J.C. and Phillips, E.J. (1993) Reduction of Uranium by Cytochrome c3 of Desulfovibrio vulgaris. Applied and Environmental Microbiology, 59, 3572-3576.
- 50. Pietzsch, K. and Babel, W. (2003) A Sulfate-Reducing Bacterium That Can Detoxify U(VI) and Obtain Energy via Nitrate Reduction. Journal of Basic Microbiology, 43, 348-361.
https://doi.org/10.1002/jobm.200390038 - 51. Boonchayaanant, B., Kitanidis, P.K. and Criddle, C.S. (2008) Growth and Cometabolic Reduction Kinetics of a Uranium- and Sulfate-Reducing Desulfovibrio/Clostridia Mixed Culture: Temperature Effects. Biotechnology and Bioengineering, 99, 1107-1119.
https://doi.org/10.1002/bit.21670 - 52. Truex, M.J., Peyton, B.M., Valentine, N.B. and Gorby, Y.A. (1997) Kinetics of U(VI) Reduction by a Dissimilatory Fe(III)-Reducing Bacterium under Non-Growth Conditions. Biotechnology and Bioengineering, 55, 490-496.
https://doi.org/10.1002/(SICI)1097-0290(19970805)55:3<490::AID-BIT4>3.0.CO;2-7 - 53. Bencheikh-Latmani, R., et al. (2005) Global Transcriptional Profiling of Shewanella oneidensis MR-1 during Cr(VI) and U(VI) Reduction. Applied and Environmental Microbiology, 71, 7453-7460.
https://doi.org/10.1128/AEM.71.11.7453-7460.2005 - 54. Wade, R. and Di Christina, T.J. (2000) Isolation of U(VI) Reduction-Deficient Mutants of Shewanella putrefaciens. FEMS Microbiology Letters, 184, 143-148.
https://doi.org/10.1111/j.1574-6968.2000.tb09005.x - 55. Cao, B., et al. (2011) Contribution of Extracellular Polymeric Substances from Shewanella sp. HRCR-1 Biofilms to U(VI) Immobilization. Environmental Science & Technology, 45, 5483-5490.
https://doi.org/10.1021/es200095j - 56. Romero-Gonzalez, M., Nwaobi, B., Hufton, J.M. and Gilmour, D.J. (2016) Ex Situ Bioremediation of U(VI) from Contaminated Mine Water Using Acidithiobacillus ferrooxidans Strains. Frontiers in Environmental Science, 4.
https://doi.org/10.3389/fenvs.2016.00039 - 57. Gerber, U., et al. (2016) Combined Use of Flow Cytometry and Microscopy to Study the Interactions between the Gram-Negative betaproteobacterium Acidovorax facilis and Uranium (VI). Journal of Hazardous Materials, 317, 127-134.
- 58. Basu, A., Sanford, R.A., Johnson, T.M., Lundstrom, C.C. and Loffler, F.E. (2014) Uranium Isotopic Fractionation Factors during U(VI) Reduction by Bacterial Isolates. Geochimica et Cosmochimica Acta, 136, 100-113.
- 59. Boyanov, M.I., et al. (2011) Solution and Microbial Controls on the Formation of Reduced U(IV) Species. Environmental Science & Technology, 45, 8336-8344.
https://doi.org/10.1021/es2014049 - 60. Rademacher, L.K., et al. (2006) Experimentally Determined Uranium Isotope Fractionation during Reduction of Hexavalent U by Bacteria and Zero Valent Iron. Environmental Science & Technology, 40, 6943-6948.
https://doi.org/10.1021/es0604360 - 61. Rui, X., et al. (2013) Bioreduction of Hydrogen Uranyl Phosphate: Mechanisms and U(IV) Products. Environmental Science & Technology, 47, 5668-5678.
https://doi.org/10.1021/es305258p - 62. Sharp, J.O., et al. (2009) Structural Similarities between Biogenic Uraninites Produced by Phylogenetically and Metabolically Diverse Bacteria. Environmental Science & Technology, 43, 8295-8301.
https://doi.org/10.1021/es901281e - 63. Akob, D.M., et al. (2008) Functional Diversity and Electron Donor Dependence of Microbial Populations Capable of U(VI) Reduction in Radionuclide-Contaminated Subsurface Sediments. Applied and Environmental Microbiology, 74, 3159-3170.
https://doi.org/10.1128/AEM.02881-07 - 64. Sánchez-Castro, I., et al. (2017) Screening of Bacterial Strains Isolated from Uranium Mill Tailings Porewaters for Bioremediation Purposes. Journal of Environmental Radioactivity, 166, 130-141.
- 65. Yao, T., et al. (2016) Biosorption of Eu(III) and U(VI) on Bacillus subtilis: Macroscopic and Modeling Investigation. Journal of Molecular Liquids, 219, 32-38.
- 66. Koribanics, N.M., et al. (2015) Spatial Distribution of an Uranium-Respiring Betaproteobacterium at the Rifle, CO Field Research Site. PLoS One, 10, e0123378.
- 67. Park, D.M. and Jiao, Y. (2014) Modulation of Medium pH by Caulobacter crescentus Facilitates Recovery from Uranium-Induced Growth Arrest. Applied and Environmental Microbiology, 80, 5680-5688.
https://doi.org/10.1128/AEM.01294-14 - 68. Yung, M.C. and Jiao, Y. (2014) Biomineralization of Uranium by PhoY Phosphatase Activity Aids Cell Survival in Caulobacter crescentus. Applied and Environmental Microbiology, 80, 4795-4804.
https://doi.org/10.1128/AEM.01050-14 - 69. Yung, M.C., et al. (2014) Shotgun Proteomic Analysis Unveils Survival and Detoxification Strategies by Caulobacter crescentus during Exposure to Uranium, Chromium, and Cadmium. Journal of Proteome Research, 13, 1833-1847.
https://doi.org/10.1021/pr400880s - 70. Yung, M.C., et al. (2015) Transposon Mutagenesis Paired with Deep Sequencing of Caulobacter crescentus under Uranium Stress Reveals Genes Essential for Detoxification and Stress Tolerance. Journal of Bacteriology, 197, 3160-3172.
https://doi.org/10.1128/JB.00382-15 - 71. Sani, R., Peyton, B., Smith, W., Apel, W. and Petersen, J. (2002) Dissimilatory Reduction of Cr(VI), Fe(III), and U(VI) by Cellulomonas Isolates. Applied Microbiology and Biotechnology, 60, 192-199.
https://doi.org/10.1007/s00253-002-1069-6 - 72. Sivaswamy, V., et al. (2011) Multiple Mechanisms of Uranium Immobilization by Cellulomonas sp. Strain ES6. Biotechnology and Bioengineering, 108, 264-276.
https://doi.org/10.1002/bit.22956 - 73. Macaskie, L.E., Bonthrone, K.M., Yong, P. and Goddard, D.T. (2000) Enzymically Mediated Bioprecipitation of Uranium by a Citrobacter sp.: A Concerted Role for Exocellular Lipopolysaccharide and Associated Phosphatase in Biomineral Formation. Microbiology, 146, 1855-1867.
https://doi.org/10.1099/00221287-146-8-1855 - 74. Lack, J.G., et al. (2002) Immobilization of Radionuclides and Heavy Metals through Anaerobic Bio-Oxidation of Fe(II). Applied and Environmental Microbiology, 68, 2704-2710.
https://doi.org/10.1128/AEM.68.6.2704-2710.2002 - 75. Sundararajan, M., Campbell, A.J. and Hillier, I.H. (2008) Catalytic Cycles for the Reduction of [UO2]2+ by Cytochrome c7 Proteins Proposed from DFT Calculations. Journal of Physical Chemistry A, 112, 4451-4457.
https://doi.org/10.1021/jp800209p - 76. Sarkar, A., Sar, P. and Islam, E. (2016) Hexavalent Chromium Reduction by Microbacterium oleivorans A1: A Possible Mechanism of Chromate Detoxification and Bioremediation. Recent Patents on Biotechnology, 9, 116-129.
https://doi.org/10.2174/187220830902160308192126 - 77. Thorgersen, M.P., et al. (2017) A Highly Expressed High-Molecular-Weight S-Layer Complex of Pelosinus sp. Strain UFO1 Binds Uranium. Applied and Environmental Microbiology, 83, e03044.
https://doi.org/10.1128/aem.03044-16 - 78. Brady, D., Stoll, A.D., Starke, L. and Duncan, J.R. (1994) Chemical and Enzymatic Extraction of Heavy Metal Binding Polymers from Isolated Cell Walls of Saccharomyces cerevisiae. Biotechnology and Bioengineering, 44, 297-302.
https://doi.org/10.1002/bit.260440307 - 79. Wang, T., Zheng, X., Wang, X., Lu, X. and Shen, Y. (2017) Different Biosorption Mechanisms of Uranium(VI) by Live and Heat-Killed Saccharomyces cerevisiae under Environmentally Relevant Conditions. Journal of Environmental Radioactivity, 167, 92-99.
- 80. Zheng, X.-Y., Wang, X.-Y., Shen, Y.-H., Lu, X. and Wang, T.-S. (2017) Biosorption and Biomineralization of Uranium(VI) by Saccharomyces cerevisiae—Crystal Formation of Chernikovite. Chemosphere, 175, 161-169.
- 81. Macaskie, L.E., et al. (2005) A Novel Non Line-of-Sight Method for Coating Hydroxyapatite onto the Surfaces of Support Materials by Biomineralization. Journal of Biotechnology, 118, 187-200.
- 82. Newsome, L., Morris, K. and Lloyd, J.R. (2015) Uranium Biominerals Precipitated by an Environmental Isolate of Serratia under Anaerobic Conditions. PLoS One, 10, e0132392.
https://doi.org/10.1371/journal.pone.0132392 - 83. Zou, L., Chen, Z., Zhang, X., Liu, P. and Li, X. (2014) Phosphate Promotes Uranium (VI) Adsorption in Staphylococcus aureus LZ-01. Letters in Applied Microbiology, 59, 528-534.
https://doi.org/10.1111/lam.12310 - 84. Friis, N. and Myers-Keith, P. (1986) Biosorption of Uranium and Lead by Streptomyces longwoodensis. Biotechnology and Bioengineering, 28, 21-28.
https://doi.org/10.1002/bit.260280105 - 85. Bengtsson, L., Johansson, B., Hackett, T.J., McHale, L. and McHale, A.P. (1995) Studies on the Biosorption of Uranium by Talaromyces emersonii CBS 814.70 Biomass. Applied Microbiology and Biotechnology, 42, 807-811.
https://doi.org/10.1007/BF00171965 - 86. Beller, H.R., et al. (2009) Identification of c-type Cytochromes Involved in Anaerobic, Bacterial U(IV) Oxidation. Biodegradation, 20, 45-53. https://doi.org/10.1007/s10532-008-9198-y
- 87. Holmes, D.E., Finneran, K.T., O’Neil, R.A. and Lovley, D.R. (2002) Enrichment of Members of the Family Geobacteraceae Associated with Stimulation of Dissimilatory Metal Reduction in Uranium-Contaminated Aquifer Sediments. Applied and Environmental Microbiology, 68, 2300-2306.
https://doi.org/10.1128/AEM.68.5.2300-2306.2002 - 88. Mohanty, S.R., et al. (2008) Biogeochemical Processes in Ethanol Stimulated Uranium-Contaminated Subsurface Sediments. Environmental Science & Technology, 42, 4384-4390.
https://doi.org/10.1021/es703082v - 89. Moon, H.S., et al. (2010) Microbial Reduction of Uranium under Iron- and Sulfate-Reducing Conditions: Effect of Amended Goethite on Microbial Community Composition and Dynamics. Water Research, 44, 4015-4028.
- 90. Pearce, C.I., et al. (2012) Pore-Scale Characterization of Biogeochemical Controls on Iron and Uranium Speciation under Flow Conditions. Environmental Science & Technology, 46, 7992-8000.
https://doi.org/10.1021/es301050h - 91. N’Guessan, A.L., Vrionis, H.A., Resch, C.T., Long, P.E. and Lovley, D.R. (2008) Sustained Removal of Uranium from Contaminated Groundwater Following Stimulation of Dissimilatory Metal Reduction. Environmental Science & Technology, 42, 2999-3004.
https://doi.org/10.1021/es071960p - 92. Renshaw, J.C., et al. (2005) Bioreduction of Uranium: Environmental Implications of a Pentavalent Intermediate. Environmental Science & Technology, 39, 5657-5660.
https://doi.org/10.1021/es048232b - 93. O’Loughlin, E.J., Kelly, S.D. and Kemner, K.M. (2010) XAFS Investigation of the Interactions of U(VI) with Secondary Mineralization Products from the Bioreduction of Fe(III) Oxides. Environmental Science & Technology, 44, 1656-1661.
https://doi.org/10.1021/es9027953 - 94. Vuorinen, A., Hiltunen, P. and Tuovinen, O.H. (1986) Redox and Precipitation Reactions of Iron and Uranium Solubilized from Ore Materials. Hydrometallurgy, 15, 297-301.
- 95. Li, J., et al. (2016) Removal of Uranium from Uranium Plant Wastewater Using Zero-Valent Iron in an Ultrasonic Field. Nuclear Engineering and Technology, 48, 744-750.
- 96. Alexandratos, V.G., Behrends, T. and Van Cappellen, P. (2017) Fate of Adsorbed U (VI) during Sulfidization of Lepidocrocite and Hematite. Environmental Science & Technology, 51, 2140-2150.
https://doi.org/10.1021/acs.est.6b05453 - 97. Shelobolina, E.S., et al. (2007) Importance of c-Type Cytochromes for U(VI) Reduction by Geobacter sulfurreducens. BMC Microbiology, 7, 16.
https://doi.org/10.1186/1471-2180-7-16 - 98. Marshall, M.J., et al. (2006) c-Type Cytochrome-Dependent Formation of U(IV) Nanoparticles by Shewanella oneidensis. PLoS Biology, 4, e268.
https://doi.org/10.1371/journal.pbio.0040268 - 99. Holmes, D.E., et al. (2008) Transcriptome of Geobacter uraniireducens Growing in Uranium-Contaminated Subsurface Sediments. The ISME Journal, 3, 216-230.
https://doi.org/10.1038/ismej.2008.89 - 100. Yun, J., Malvankar, N.S., Ueki, T. and Lovley, D.R. (2016) Functional Environmental Proteomics: Elucidating the Role of a c-Type Cytochrome Abundant during Uranium Bioremediation. The ISME Journal, 10, 310-320.
https://doi.org/10.1038/ismej.2015.113 - 101. Shi, L., et al. (2011) Identification and Characterization of UndAHRCR-6, an Outer Membrane Endecaheme c-Type Cytochrome of Shewanella sp. Strain HRCR-6. Applied and Environmental Microbiology, 77, 5521-5523.
https://doi.org/10.1128/AEM.00614-11 - 102. Payne, R., et al. (2004) Interaction between Uranium and the Cytochrome c3 of Desulfovibrio desulfuricans Strain G20. Archives of Microbiology, 181, 398-406.
https://doi.org/10.1007/s00203-004-0671-7 - 103. Nikaido, H. (2003) Molecular Basis of Bacterial Outer Membrane Permeability Revisited. Microbiology and Molecular Biology Reviews, 67, 593-656.
https://doi.org/10.1128/MMBR.67.4.593-656.2003 - 104. Cologgi, D.L., Lampa-Pastirk, S., Speers, A.M., Kelly, S.D. and Reguera, G. (2011) Extracellular Reduction of Uranium via Geobacter Conductive Pili as a Protective Cellular Mechanism. Proceedings of the National Academy of Sciences, 108, 15248-15252.
https://doi.org/10.1073/pnas.1108616108 - 105. Shi, L., et al. (2016) Extracellular Electron Transfer Mechanisms between Microorganisms and Minerals. Nature Reviews Microbiology, 14, 651-662.
https://doi.org/10.1038/nrmicro.2016.93 - 106. Lampa-Pastirk, S., et al. (2016) Thermally Activated Charge Transport in Microbial Protein Nanowires. Scientific Reports, 6, Article Number: 23517.
https://doi.org/10.1038/srep23517 - 107. Orellana, R., et al. (2013) U(VI) Reduction by Diverse Outer Surface c-Type Cytochromes of Geobacter sulfurreducens. Applied and Environmental Microbiology, 79, 6369-6374.
https://doi.org/10.1128/AEM.02551-13 - 108. Cologgi, D.L., Speers, A.M., Bullard, B.A., Kelly, S.D. and Reguera, G. (2014) Enhanced Uranium Immobilization and Reduction by Geobacter sulfurreducens Biofilms. Applied and Environmental Microbiology, 80, 6638-6646.
https://doi.org/10.1128/AEM.02289-14 - 109. Steidl, R.J., Lampa-Pastirk, S. and Reguera, G. (2016) Mechanistic Stratification in Electroactive Biofilms of Geobacter sulfurreducens Mediated by Pilus Nanowires. Nature Communications, 7, Article Number: 12217.
https://doi.org/10.1038/ncomms12217 - 110. Li, X., Zhang, H., Ma, Y., Liu, P. and Krumholz, L.R. (2014) Genes Required for Alleviation of Uranium Toxicity in Sulfate Reducing Bacterium Desulfovibrio alaskensis G20. Ecotoxicology, 23, 726-733.
- 111. Mtimunye, P.J. and Chirwa, E.M. (2014) Characterization of the Biochemical-Pathway of Uranium (VI) Reduction in Facultative Anaerobic Bacteria. Chemosphere, 113, 22-29.
- 112. Li, X. and Krumholz, L.R. (2009) Thioredoxin Is Involved in U(VI) and Cr(VI) Reduction in Desulfovibrio desulfuricans G20. Journal of Bacteriology, 191, 4924-4933.
https://doi.org/10.1128/JB.00197-09 - 113. Bargar, J.R., et al. (2013) Uranium Redox Transition Pathways in Acetate-Amended Sediments. Proceedings of the National Academy of Sciences, 110, 4506-4511.
https://doi.org/10.1073/pnas.1219198110 - 114. Stylo, M., Neubert, N., Roebbert, Y., Weyer, S. and Bernier-Latmani, R. (2015) Mechanism of Uranium Reduction and Immobilization in Desulfovibrio vulgaris Biofilms. Environmental Science & Technology, 49, 10553-10561.
https://doi.org/10.1021/acs.est.5b01769 - 115. Baiget, M., Constanti, M., Lopez, M.T. and Medina, F. (2013) Uranium Removal from a Contaminated Effluent Using a Combined Microbial and Nanoparticle System. New Biotechnology, 30, 788-792.
- 116. Suriya, J., et al. (2016) Assessment of Bacterial Community Composition in Response to Uranium Levels in Sediment Samples of Sacred Cauvery River. Applied Microbiology and Biotechnology, 1-11.
- 117. Abdelouas, A., Lutze, W. and Nuttall, E. (1998) Chemical Reactions of Uranium in Ground Water at a Mill Tailings Site. Journal of Contaminant Hydrology, 34, 343-361.
- 118. Liu, C., Jeon, B.H., Zachara, J.M. and Wang, Z. (2007) Influence of Calcium on Microbial Reduction of Solid Phase Uranium(VI). Biotechnology and Bioengineering, 97, 1415-1422.
https://doi.org/10.1002/bit.21357 - 119. Bernier-Latmani, R., et al. (2010) Non-Uraninite Products of Microbial U(VI) Reduction. Environmental Science & Technology, 44, 9456-9462.
https://doi.org/10.1021/es101675a - 120. Alessi, D.S., et al. (2014) Speciation and Reactivity of Uranium Products Formed during in Situ Bioremediation in a Shallow Alluvial Aquifer. Environmental Science & Technology, 48, 12842-12850.
https://doi.org/10.1021/es502701u - 121. Alessi, D.S., et al. (2012) Quantitative Separation of Monomeric U(IV) from UO2 in Products of U(VI) Reduction. Environmental Science & Technology, 46, 6150-6157.
https://doi.org/10.1021/es204123z - 122. Ganesh, R., Robinson, K.G., Reed, G.D. and Sayler, G.S. (1997) Reduction of Hexavalent Uranium from Organic Complexes by Sulfate- and Iron-Reducing Bacteria. Applied Microbiology and Biotechnology, 63, 4385-4391.
- 123. Jeon, O.H., et al. (2004) Microbial Reduction of U(VI) at the Solid-Water Interface. Environmental Science & Technology, 38, 5649-5655.
https://doi.org/10.1021/es0496120 - 124. N’Guessan, A.L., et al. (2010) Molecular Analysis of Phosphate Limitation in Geobacteraceae during the Bioremediation of a Uranium-Contaminated Aquifer. The ISME Journal, 4, 253-266.
https://doi.org/10.1038/ismej.2009.115 - 125. Neiss, J., Stewart, B.D., Nico, P.S. and Fendorf, S. (2007) Speciation-Dependent Microbial Reduction of Uranium within Iron-Coated Sands. Environmental Science & Technology, 41, 7343-7348.
https://doi.org/10.1021/es0706697 - 126. Sheng, L. and Fein, J.B. (2014) Uranium Reduction by Shewanella oneidensis MR-1 as a Function of NaHCO3 Concentration: Surface Complexation Control of Reduction Kinetics. Environmental Science & Technology, 48, 3768-3775.
https://doi.org/10.1021/es5003692 - 127. Stewart, B.D., Amos, R.T. and Fendorf, S. (2011) Effect of Uranium(VI) Speciation on Simultaneous Microbial Reduction of Uranium(VI) and Iron(III). Journal of Environmental Quality, 40, 90-97.
https://doi.org/10.2134/jeq2010.0304 - 128. Stewart, B.D., Neiss, J. and Fendorf, S. (2007) Quantifying Constraints Imposed by Calcium and Iron on Bacterial Reduction of Uranium(VI). Journal of Environmental Quality, 36, 363-372.
https://doi.org/10.2134/jeq2006.0058 - 129. Gao, W. and Francis, A.J. (2013) Fermentation and Hydrogen Metabolism Affect Uranium Reduction by Clostridia. ISRN Biotechnology, 2013, 11 p.
https://doi.org/10.5402/2013/657160 - 130. Liu, C., Gorby, Y.A., Zachara, J.M., Fredrickson, J.K. and Brown, C.F. (2002) Reduction Kinetics of Fe(III), Co(III), U(VI), Cr(VI), and Tc(VII) in Cultures of Dissimilatory Metal-Reducing Bacteria. Biotechnology and Bioengineering, 80, 637-649.
https://doi.org/10.1002/bit.10430 - 131. Liu, C., Zachara, J.M., Fredrickson, J.K., Kennedy, D.W. and Dohnalkova, A. (2002) Modeling the Inhibition of the Bacteral Reduction of U(VI) by Beta-MnO2(s). Environmental Science & Technology, 36, 1452-1459.
https://doi.org/10.1021/es011159u - 132. Liu, J.-X., et al. (2015) U(VI) Reduction by Shewanella oneidensis Mediated by Anthraquinone-2-Sulfonate. Transactions of Nonferrous Metals Society of China, 25, 4144-4150.
- 133. Luan, F., Gorski, C.A. and Burgos, W.D. (2014) Thermodynamic Controls on the Microbial Reduction of Iron-Bearing Nontronite and Uranium. Environmental Science & Technology, 48, 2750-2758.
https://doi.org/10.1021/es404885e - 134. Suzuki, Y., Kitatsuji, Y., Ohnuki, T. and Tsujimura, S. (2010) Flavin Mononucleotide Mediated Electron Pathway for Microbial U(VI) Reduction. Physical Chemistry Chemical Physics, 12, 10081-10087.
https://doi.org/10.1039/c0cp00339e - 135. Mosher, J.J., et al. (2012) Microbial Community Succession during Lactate Amendment and Electron Acceptor Limitation Reveals a Predominance of Metal-Reducing Pelosinus spp. Applied Microbiology and Biotechnology, 78, 2082-2091.
https://doi.org/10.1128/AEM.07165-11 - 136. Istok, J.D., et al. (2004) In Situ Bioreduction of Technetium and Uranium in a Nitrate-Contaminated Aquifer. Environmental Science & Technology, 38, 468-475.
https://doi.org/10.1021/es034639p - 137. Brodie, E.L., et al. (2006) Application of a High-Density Oligonucleotide Microarray Approach to Study Bacterial Population Dynamics during Uranium Reduction and Reoxidation. Applied Microbiology and Biotechnology, 72, 6288-6298.
https://doi.org/10.1128/AEM.00246-06 - 138. Wagner, D.D., et al. (2012) Genomic Determinants of Organohalide-Respiration in Geobacter lovleyi, an Unusual Member of the Geobacteraceae. BMC Genomics, 13, 200.
https://doi.org/10.1186/1471-2164-13-200 - 139. Salome, K.R., Beazley, M.J., Webb, S.M., Sobecky, P.A. and Taillefert, M. (2017) Biomineralization of U(VI) Phosphate Promoted by Microbially-Mediated Phytate Hydrolysis in Contaminated Soils. Geochimica et Cosmochimica Acta, 197, 27-42.
- 140. Orellana, R., et al. (2014) Proteome of Geobacter sulfurreducens in the Presence of U(VI). Microbiology, 160, 2607-2617.
https://doi.org/10.1099/mic.0.081398-0 - 141. Giloteaux, L., et al. (2013) Characterization and Transcription of Arsenic Respiration and Resistance Genes during in Situ Uranium Bioremediation. The ISME Journal, 7, 370-383.
https://doi.org/10.1038/ismej.2012.109 - 142. Parks, J.M., et al. (2013) The Genetic Basis for Bacterial Mercury Methylation. Science, 339, 1332-1335.
https://doi.org/10.1126/science.1230667 - 143. Gillan, D.C. (2016) Metal Resistance Systems in Cultivated Bacteria: Are They Found in Complex Communities? Current Opinion in Biotechnology, 38, 123-130.
https://doi.org/10.1016/j.copbio.2016.01.012 - 144. Panak, P., et al. (2000) Complex Formation of U(VI) with Bacillus-Isolates from a Uranium Mining Waste Pile. Radiochimica Acta, 88, 71-76.
https://doi.org/10.1524/ract.2000.88.2.071 - 145. Handley-Sidhu, S., et al. (2014) Bacterially Produced Calcium Phosphate Nanobiominerals: Sorption Capacity, Site Preferences, and Stability of Captured Radionuclides. Environmental Science & Technology, 48, 6891-6898.
https://doi.org/10.1021/es500734n - 146. Koster van Groos, P.G., et al. (2016) Uranium Fate in Wetland Mesocosms: Effects of Plants at Two Iron Loadings with Different pH Values. Chemosphere, 163, 116-124.
https://doi.org/10.1016/j.chemosphere.2016.08.012 - 147. Nie, X., et al. (2016) Removel of Uranium from Aqueous Solutions by Spirodela Punctata as the Mechanism of Biomineralization. Procedia Environmental Sciences, 31, 382-391.
https://doi.org/10.1016/j.proenv.2016.02.060 - 148. Dang, D.H., Novotnik, B., Wang, W., Georg, R.B. and Evans, R.D. (2016) Uranium Isotope Fractionation during Adsorption, (Co)Precipitation and Biotic Reduction. Environmental Science & Technology.
- 149. Stylo, M., et al. (2015) Uranium Isotopes Fingerprint Biotic Reduction. Proceedings of the National Academy of Sciences, 112, 5619-5624.
https://doi.org/10.1073/pnas.1421841112 - 150. Wilkins, M.J., et al. (2011) Development of a Biomarker for Geobacter Activity and Strain Composition; Proteogenomic Analysis of the Citrate Synthase Protein during Bioremediation of U(VI). Microbial Biotechnology, 4, 55-63.
https://doi.org/10.1111/j.1751-7915.2010.00194.x - 151. Smith, M.B., et al. (2015) Natural Bacterial Communities Serve as Quantitative Geochemical Biosensors. MBio, 6, e00326-e00315.
https://doi.org/10.1128/mbio.00326-15 - 152. Zhu, X., Kriegel, A.M., Boustany, C.A. and Blake, D.A. (2011) Single-Chain Variable Fragment (scFv) Antibodies Optimized for Environmental Analysis of Uranium. Analytical Chemistry, 83, 3717-3724.
https://doi.org/10.1021/ac200159x - 153. Melton, S.J., et al. (2009) Field-Based Detection and Monitoring of Uranium in Contaminated Groundwater Using Two Immuno Sensors. Environmental Science & Technology, 43, 6703-6709.
https://doi.org/10.1021/es9007239 - 154. Zhang, H., et al. (2015) A Turn-Off Fluorescent Biosensor for the Rapid and Sensitive Detection of Uranyl Ion Based on Molybdenum Disulfide Nanosheets and Specific DNAzyme. Spectrochim Acta A Mol Biomol Spectrosc, 146, 1-6.
- 155. Sani, R.K., Peyton, B.M. and Dohnalkova, A. (2008) Comparison of Uranium(VI) Removal by Shewanella oneidensis MR-1 in Flow and Batch Reactors. Water Research, 42, 2993-3002.
- 156. Chang, H.S., et al. (2014) Uranium Immobilization in an Iron-Rich Rhizosphere of a Native Wetland Plant from the Savannah River Site under Reducing Conditions. Environmental Science & Technology, 48, 9270-9278.
https://doi.org/10.1021/es5015136 - 157. Ayala-Parra, P., Sierra-Alvarez, R. and Field, J.A. (2016) Algae as an Electron Donor Promoting Sulfate Reduction for the Bioremediation of Acid Rock Drainage. Journal of Hazardous Materials, 317, 335-343.
- 158. Arnold, T., Großmann, K. and Baumann, N. (2009) Uranium Speciation in Biofilms Studied by Laser Fluorescence Techniques. Analytical and Bioanalytical Chemistry, 396, 1641-1653.
https://doi.org/10.1007/s00216-009-3296-5 - 159. Jiang, S. and Hur, H.G. (2013) Effects of the Anaerobic Respiration of Shewanella oneidensis MR-1 on the Stability of Extracellular U(VI) Nanofibers. Microbes and Environments, 28, 312-315.
- 160. Jiang, S., et al. (2011) Bacterial Formation of Extracellular U(VI) Nanowires. Chemical Communications, 47, 8076-8078.
https://doi.org/10.1039/c1cc12554k - 161. Schofield, E.J., et al. (2008) Structure of Biogenic Uraninite Produced by Shewanella oneidensis Strain MR-1. Environmental Science & Technology, 42, 7898-7904.
https://doi.org/10.1021/es800579g





