Advances in Microbiology
Vol. 2 No. 2 (2012) , Article ID: 19688 , 9 pages DOI:10.4236/aim.2012.22021
Evaluation of Real-Time PCR and Culturing for the Detection of Leptospires in Canine Samples
1WHO/FAO/OIE and National Collaborating Centre for Reference and Research on Leptospirosis, Department of Biomedical Research, Royal Tropical Institute, Amsterdam, The Netherlands
2MSD Animal Health, Department Microbiological Research and Development, Boxmeer, The Netherlands
Email: *a.ahmed@kit.nl
Received February 21, 2012; revised March 28, 2012; accepted April 5, 2012
Keywords: Leptospirosis; Diagnosis; PCR; Evaluation; Canine; Experimental infection; Challenge; Infection; Kinetics
ABSTRACT
Validated real-time PCRs detecting leptospires for veterinary purposes are not readily available. This paper describes the prospective evaluation of a SYBR Green-based real-time PCR on serum samples collected from experimentally infected dogs. Compared to culturing, the assay had a diagnostic sensitivity and specificity of 91.7% and 90.0%, respectively. Culturing for part is complementary to PCR and preferably both should be applied for diagnosis and vaccine challenge experiments. In a subsequent prospective study on the dynamics of experimental infections with serovars Canicola and Copenhageni and serovar complex Bananal-Liangguang in young and adult dogs, the PCR was applied on serum samples in conjunction with culturing on blood, urine and kidney samples with the following results: 1) Leptospires persisted longer in the blood of young dogs than of adult ones; 2) Numbers of viable leptospires in the blood are rapidly reduced but DNA remains occasionally detectable up to 7 days post infection; 3) Appearance of viable leptospires in the urine follows a biphasic dynamics; 4) EDTA hampers effective culturing from blood samples; 5) Serovar Canicola persists longer in the blood of dogs than Copenhageni and Bananal-Liangguang. Together with a markedly higher recovery rate from kidney samples (71% compared to respectively 33% and 0% in young dogs) this probably reflects the adaptation power of Canicola to its canine maintenance host. Appearance of leptospires in the urine samples indicates that experimental infections have been successful. PCR presents a valuable adjunct to the diagnosis of veterinary leptospirosis and the follow-up in vaccine protection experiments.
1. Introduction
Leptospires are spirochetes of the genus Leptospira that are categorized into three groups comprising saprophytic, intermediate, and pathogenic species. Pathogenic leptospires are the causative agent of leptospirosis, which is a worldwide zoonotic disease with a significant impact on both public and veterinary health. The pathogen is maintained in the renal tubules and genital tract of many domestic and wild mammalian species [1]. Notably small mammals are considered as main maintenance hosts. The maintenance host excretes the pathogen with the urine into the environment where leptospires can survive up to several months depending on favourable humid and warm conditions [2]. Accidental hosts acquire the infection via broken skin or mucous membrane by direct contact with infected urine or indirect contact through contaminated soil, food or water [3,4]. The serovar is the basic taxon of Leptospira, characterized by serological criteria determined by the Cross Agglutinin Absorption Test (CAAT). To date, about 250 pathogenic serovars have been identified [4-7]. At present leptospirosis is considered a globally re-emerging disease with frequent outbreaks on all continents, often but not exclusively due to disasters [8-10] and sporting events [11,12].
Leptospirosis is a protean disease in humans and animals that is often not recognized or confused with other diseases. Leptospirosis in animals shows a variety of clinical signs and symptoms much similar to humans, albeit that details may differ between different animal species. Leptospirosis in pigs is mainly associated with reproductive failure. Disease severity in cattle might depend on the infecting serovar, the vaccination status, and the age. Adult cattle with a recent infection with serovar Hardjo may show transient effects such as abnormal milk production but abortion and stillbirth might occur in chronic infections [13]. Acute leptospirosis might be severe in calves. Typically calves show fever, anorexia, dyspnea, icterus, hemoglobinuria, and haemolytic anemia. In dogs serovars Canicola and Icterohaemorrhagiae/Copenhageni are the typical causative serovars, although several additional serovars, including Grippotyphosa, have been implicated [1,14,15]. The incubation period of the disease can be a few days. As with cattle, the severity of clinical signs is influenced by age, vaccination status, the inherent virulence of a particular serovar and the route and degree of exposure. In peracute to subacute disease, dogs may die without clinical signs. Early clinical signs may include fever, accompanied by shivering, generalized muscle tenderness and reluctance to move. The clinical signs of acute renal failure include polyuria, polydipsia, dehydration, vomiting, diarrhea, inappetence, lethargy, abdominal pain or some combination of these signs. Oliguria or anuria also may occur. Other reported manifestations of infection include conjunctivitis, uveitis and tachypnea or dyspnea because of acute respiratory distress syndrome or leptospiral pulmonary hemorrhage syndrome (LPHS). Icterus may present with other signs of hepatic failure [16]. For laboratory confirmation, the microscopic agglutination test (MAT) is the standard diagnostic test detecting agglutinating anti-Leptospira antibodies as a sign of infection. MAT can be applied on serum samples from virtually all mammalian species. While serology is a suitable test to detect acute infection, culturing from urine samples is more appropriate for chronic infections in which serology often is negative or inconclusive [17]. However, whereas isolation of leptospires provides proof of infection, culturing may take months and is too slow to assess a current situation. This bottleneck also hampers potency testing of vaccines in animals, where sterilizing immunity is mostly recorded by a reduced circulation of leptospires in the blood and the absence of colonization of kidneys. The polymerase chain reaction (PCR) is a valuable adjunct to the detection of leptospires in suspected patients or chronic carriers [18-22]. To date, three real-time PCR schemes have been well validated for the use in the diagnosis of human leptospirosis [21,23]. However, their value for animal investigations has not been assessed. One of these realtime PCRs uses the SYBR Green chemistry and targets the secY gene [21]. This PCR has a particular advantage in that the secY gene is highly discriminative in phylogenetic characterization providing an excellent target for molecular epidemiological investigations [24]. In a previous paper, we described the diagnostic accuracy of this real-time PCR on clinical samples from human patients using both MAT and culture as reference [21]. In this paper we investigate the applicability of this realtime PCR to follow up experimental infections in dogs, using culture as the reference test to define infection. Subsequently, both PCR and culture were applied on blood and urine samples from groups of young and adult dogs experimentally infected with serovars Copenhageni, Canicola and Bananal-Liangguang 11808 to investigate infection dynamics.
2. Materials and Methods
2.1. Strains and Culturing
Strains used are Leptospira interrogans, serovar Copenhageni (serogroup Icterohaemorrhagiae), strain CF1 and serovar Canicola (serogroup Canicola), strain Moulton, and L. kirschneri, serovar complex Bananal/Liangguang (serogroup Grippotyphosa), in this paper denoted as SvC Bananal-Liangguang, strain 11808 from the collection of MSD Animal Health, Boxmeer, The Netherlands. Culturing was performed in liquid Ellinghausen-McCullough-Johnson-Harris (EMJH) medium as described previously [25]. Standard procedures were followed for inoculation of EMJH culture medium with canine specimens [2].
2.2. The Study Design
Experimental infection and culturing was performed at MSD Animal Health, Boxmeer. 200 µl portions of coded, double blinded serum samples were sent to the WHO/ FAO/OIE and National Leptospirosis Centre (NRL), KIT Biomedical Research, Amsterdam, The Netherlands for prospective testing by real-time PCR. Thus, testing of samples by culture and by real-time PCR was done independently by two distinct groups of experienced staff at MSD Animal Health and at NRL without prior knowledge of results obtained at the partner. Unless otherwise stated, for determination of the clinical sensitivity and specificity of the real-time PCR as index test, culturing was used as the reference standard to define an infection.
1) Experimental infections. For all dogs the challenge route was intraperitoneal injection (2 ml) and conjunctival instillation (0.25 ml into ventral conjunctival sac of each eye). Dogs included in this study were healthy and free of anti-Leptospira antibodies that might affect the outcome of the experiments. The bacterial cell counts (total direct microscopic count) in the several challenge materials were as follows: strain CF1, 1.0 × 109 cells per ml; strain Moulton, 5.0 × 108 (study A) or 1.0 × 109 (study B) cells per ml; strain 11808, 1.0 × 109 cells per ml. One ml blood and 5 ml urine samples were taken by intravenous puncturing and sterile puncturing of the bladder, respectively. Ethical clearance for these experiments was obtained from the Animal Experiment Ethical Committee, Nijmegen, The Netherlands.
2) Clinical Samples. Blood and sera from experimentally infected dogs were investigated in two separate experiments. The first experiment, referred to as study A, was used to validate the real-time PCR against culturing as the reference test. Study A comprised two groups of eight young dogs (13 weeks) each, infected with either serovar Copenhageni, strain CF1, or serovar Canicola, strain Moulton (Table 1). Heparin blood was collected for culturing and serum for real-time PCR on day “0” prior to inoculation and subsequently on days post infection (DPI) 3, 7, 14 and 21. On DPI 10 three dogs died, all infected with Canicola. For the determination of the diagnostic accuracy of the PCR, the validation scheme for diagnostic PCR as described by the World Organization for Animal Health was followed [5].
The second experiment, referred to as study B, comprised three groups of young dogs (13 weeks), experimentally infected with either serovar Copenhageni, strain CF1 (seven dogs), or with serovar Canicola, strain Moulton (eight dogs), or with SvC Bananal-Liangguang, strain 11808 (eight dogs). One dog in the Canicola-infected group died on DPI 14. Furthermore this experiment comprised two groups of 9 adult dogs (14 months), experimentally infected with either strain CF1 or strain Moulton (Table 2). EDTA-blood was collected on DPI 0, 1, 2, 3, 4, 7, 10, 14, and 21 for culturing. Serum samples from DPI 0, 3, and 7 were analyzed by the real-time PCR. Culturing also included urine samples collected at DPI 0, 3, 7, 14, 21 and 28 and kidney samples collected upon euthanization of the animals at DPI 28.
2.3. DNA Extraction
Leptospira DNA was extracted from 200 µl serum samples, purified, and eluted in 0.1xTE buffer pH 8.0 according to the instructions of the QIAamp DNA extraction kit (Qiagen, GmbH, D-40724 Hilden, Germany).
2.4. Real-Time PCR
SYBR Green real-time PCR was applied in this study as described previously [21]. We further refer to this realtime PCR in the text as “PCR”. PCR was performed in triplicate. Two or three positive reactions in the triplicate were scored as positive. When one out of three reactions was positive, the test was repeated in triplicate. One or more positive reactions in the repeated triplicate were included as positive results [21]. The bacterial DNA load was calculated with a standard curve of L. interrogans DNA using 10-fold dilution as described in our previous study [21].
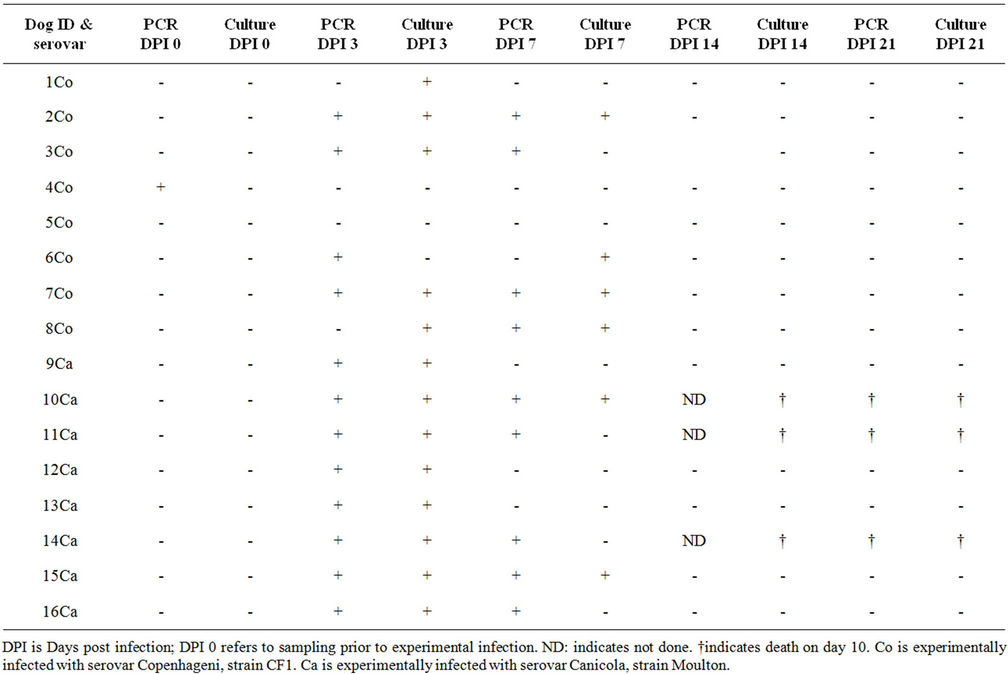
Table 1. Study A: Culturing and PCR results on blood samples from dogs experimentally infected with serovars Copenhageni and Canicola.
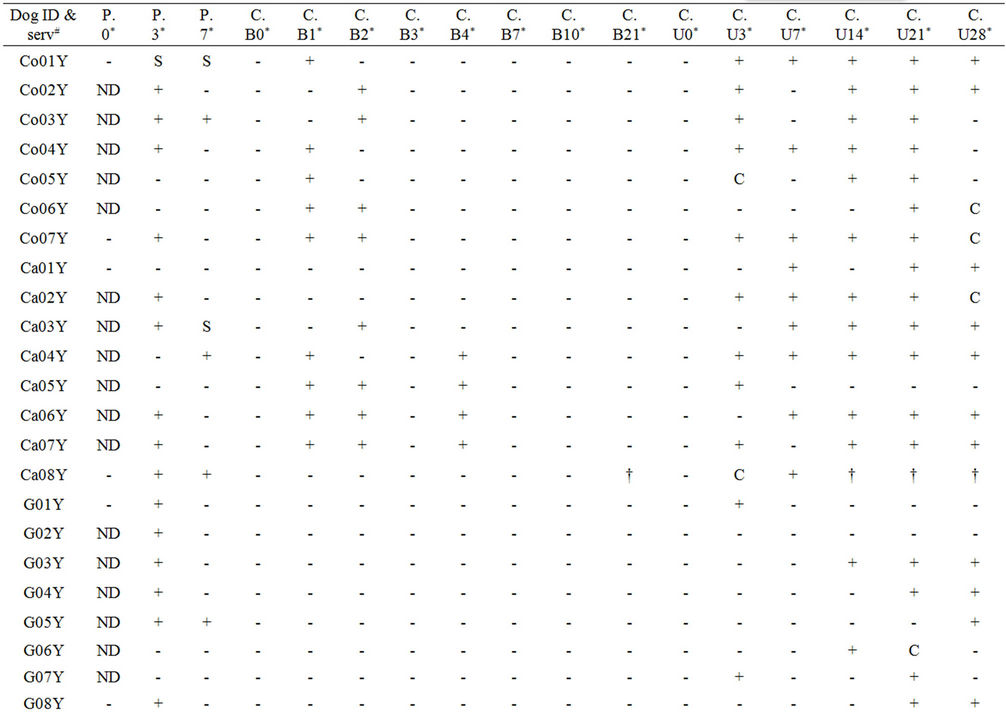

Table 2. Study B: Complete overview of PCR and culture results using blood and urine samples from dogs post infection with serovars Copenhageni, Canicola and SvC Bananal-Liangguang.
2.5. Statistics
Results were placed in two-by-two tables to calculate sensitivity, specificity and predictive values. Sensitivity was calculated as the number of true positive results divided by the sum of true positive and false negative results. Specificity was calculated as the number of true negative results divided by the sum of true negative and false positive results. Chi square test was performed to determine significance, as defined by a p value ≤ 0.05.
3. Results and Discussion
3.1. Diagnostic Accuracy of the Real-Time PCR (Study A)
In contrast to real-time PCRs for human diagnosis, validated real-time PCRs for veterinary use are scarcely available [26]. We determined the veterinary applicability of a SYBR Green-based PCR previously validated on human samples [21] using serum samples from dogs experimentally infected with either serovar Copenhageni or Canicola. Concomitant heparinized blood samples were tested for growth of leptospires in a separate experiment and results of these prospective experiments were compared (Table 1). All cultures and PCRs on serum samples collected after DPI 7 were negative, indicating the rapid reduction of the number of leptospires in the blood beyond the detection thresholds of both tests. To obtain a meaningful impression of the performance of both culture and PCR, taking the dynamics of bacteraemia into account, we calculated sensitivity (SeI) and specificity (SpI) on results of DPI 0 (no infection) and DPI 3 (infection), using experimental infection as the reference standard. PCR had a SeI of 75% (12/16) and culture had a SeI of 81.3% (13/16). The overall SeI of the two tests together is 87.5% (14/16). It should be noted that the relatively low SeI of both tests might be biased because of early clearance of leptospires from the blood of the infected dogs. The SpI for culturing was 100% since none of the samples taken at DPI 0 had a positive culture. The SpI for the PCR was 93.8% because of an unexplained positive result on a single sample collected prior to experimental infection. Mislabeling of samples, a sample switch, or cross-contamination prior to PCR execution present an obvious explanation for this discrepant result. However, original material was not available to confirm this possibility. At DPI 7, the percentage of positive PCRs declined to 56.3% and that of culturing to 37.5%.
When results were separated according to the infecting serovar, PCR and culture had a SeI of 100% (CI = 72.2% - 100%) and SpI of 87.5% (CI = 69.0% - 95.7%) for Canicola and a SeI of 75% (CI = 40.9% - 92.9%) and SpI of 90.6% (CI = 75.8% - 96.8%) for Copenhageni. Remarkably, two of the eight dogs infected with Copenhageni yielded a negative culture and PCR in all follow-up samples. An explanation for this observation could be a short survival capacity of Copenhageni in the blood of healthy dogs, since negative blood cultures on DPI 3 have been observed in comparable dog studies elsewhere [27,28]. The overall sensitivity and specificity of the PCR compared to culture was 88.9% (CI = 67.2 - 96.9) and 89.3% (CI = 78.5 - 95.5), respectively. However, these data might have lost correlation between culturing and PCR because bacterial loads are strongly reduced at late DPIs. Therefore, we argue that sensitivity and specificity of PCR is better reflected by the data from DPI 0 and 3, i.e. 91.7% (CI = 64.6 - 98.5) and 90.0% (CI = 69.9 - 97.2), respectively. We refer to these as the veterinarydiagnostic sensitivity and specificity of the PCR. It should be noted that two samples of DPI 3 were positive for culture and negative for PCR (samples 1Co and 8Co) and one sample (6Co) was culture negative and PCR positive. These differences probably have a stochastic cause in samples with a low concentration of leptospires, mostly favoring a positive culture since inoculums of about 500 µl in culture are more likely to contain leptospires than the 10 µl samples used in the PCR [21]. On the other hand, PCR can be positive on DNA from killed bacteria, explaining a positive PCR combined with a negative culture. Thus, culturing and PCR are complementary to some extent (see also Table 1) and hence application of both for the diagnosis of early acute canine leptospirosis as well as in vaccine efficacy studies should be considered for obtaining optimal results.
3.2. Prospective Study on Canine Samples by PCR and Culture (Study B)
Subsequently, PCR was applied on serum samples from study B collected at DPI 3 and 7 (Table 2). DPI 0 was only tested on some samples to confirm expected negative scores. The PCR was positive in 37.5% (15/40) and 10.3% (4/39) of the serum samples collected at DPI 3 and 7, respectively. In concordance, quantitation by PCR revealed a significant reduction in the concentration of leptospires on DPI 7 compared to DPI 3 (data not shown). Culture, which was done on collected Na-EDTA blood, was negative in all samples. Thus, it is not possible to determine sensitivity and specificity of the PCR versus culturing in this test sample. However, Canicola infections in young dogs yielded 4 positive cultures on DPI 4. Hence, these cases can be expected to have contained leptospires also on day 3. When using this subsample on DPI 3, the sensitivity and specificity of PCR compared to culturing is only 50% (CI = 15.0% - 85%) and 25% (CI = 4.6% - 70.0%), respectively.
This discrepancy with the veterinary-diagnostic sensitivity and specificity of the test as determined in study A can be explained as follows. In study A, culturing has been done on heparinized blood samples and PCR on corresponding serum samples. In contrast, in study B culturing has been done on EDTA blood samples. Omission of heparinized blood samples was attempted to investigate whether the burden of the experimental animals could be reduced by avoiding unnecessary sampling. However, this experiment showed that EDTA hampers the viability of leptospires in subsequent in vitro culturing. This has been found by others [29] and was further substantiated in a separate experiment in which we found that growth of reference strains was severely affected in EMJH culture medium supplemented by EDTA (data not shown). Hence, we conclude that this study set up does not allow correlating PCR with culturing. To meet with animal welfare as well as optimal test results, future studies will aim at the application of both culture and PCR on full blood or serum as these types of samples are suitable for either test [2,21].
3.3. Serovar-Associated Susceptibility of Infection in Young versus Adult Dogs
When splitting up the results according to infecting serovar, PCR positivity rates were more similar for each serovar, i.e. for young dogs infected with serovar Copenhageni, the PCR on DPI 3 and 7 was positive in 57.1% (4/7) (CI = 25.1% - 84.2%), and 14.3% (1/7) (CI = 2.6% - 51.3%) of the samples, respectively. For Canicola infections this was 62.5% (5/8) (CI = 30.6% - 86.3%) and 25% (2/8) (CI = 7.2% - 59.1%), respectively and for Bananal-Liangguang infections this was 75% (6/8) (CI = 41% - 93%) and 12.5% (1/8) (CI = 2.2% - 47.1%), respectively. In the category adult dogs, none of the samples were positive by PCR for the tested serovars Copenhageni and Canicola. It is therefore concluded that leptospires persist longer in the blood of young dogs than in adult ones, in concordance with several previous studies [25,30].
3.4. Investigating Dynamics of Infection by Culturing
Dynamics of leptospires in blood, urine and other body fluids or tissues after natural infection are well addressed in the literature [2,4], but surprisingly is scarcely described for experimental infection, notably on dogs [31- 33]. The performance of culturing on EDTA blood samples taken at time points up to DPI 21 and on urine samples collected until DPI 28 as listed in Table 2 enabled us to investigate the short term kinetics of experimental infections in healthy dogs. Although percentages of culture positive samples from blood samples might be underrated because of in vitro growth inhibition by the EDTA, this is not likely to affect the trends of the spirochetemic dynamics.
Consistent with PCR results, viable leptospires appeared to remain longer detectable in the blood of young animals than in adult ones. For young dogs, the Copenhageni culture was positive in 5 of 7 samples on DPI 1. However, the other two samples were positive at DPI 2, so we assume that all samples (100%) contained viable leptospires at DPI 1. This was reduced to 57.1% on DPI 2 and was negative from DPI 3 on. Canicola seemed to persist longer in detectable numbers. On DPI 1, cultures were positive in five of eight samples (62.5%), including a sample that became positive on DPI 2. This was four of eight (50%) on DPI 2 and DPI 4 (same samples as on DPI 1) while all samples were culture negative on DPI 3, probably for stochastic reasons explained above, and > DPI 4. None of the Bananal-Liangguang bacteria could be cultured from the blood, suggesting that these are either rapidly killed in the host or have a higher sensitivity to in vitro growth inhibition by EDTA. The latter possibility is more likely as this is consistent with positive PCR results up to DPI 3, suggesting some persistence of viable leptospires in the blood. Moreover, reduction of numbers of pathogenic leptospires by killing in the blood of healthy hosts is unlikely but depends on their virulence [34]. A lower virulence of Bananal-Liangguang in dogs is well possible. While this might explain the rapid clearance of viable Bananal-Liangguang from the blood, early disappearance of serovars Canicola and Copenhageni in the blood indicate other concomitant causes of reducing Leptospira concentrations, such as effective invasion of various organs leading to potentially fatal damage.
In adult animals, culture on EDTA blood samples was positive in 25% (2/8) and 12.5% (1/8) of the samples on DPI 1 and 2, respectively for Copenhageni, and 33.3% (3/9) and 22.2% (2/9), respectively for Canicola. Positive cultures on DPI 2 were also positive for DPI 1. All EDTA blood samples from adult dogs were negative from DPI 3 on.
Urine culture results in part complemented those of blood cultures. Plotting the positivity rates revealed a remarkable trend (Figure 1). Irrespective the group of dogs and the infecting serovar, all culture results showed a biphasic character (Figure 1). Colonization of the urinary tract is not expected to occur within the first week after infection. Hence the first peak is probably due to leakage or, more probably, early migration of leptospires from the glomeruli into the urinary tract [31,33], while the second peak reflects the interstitial migration of leptospires into the tubules and subsequent adherence of leptospires in the lumen of the kidneys [31,32,35]. The lowest point of the curves was found between DPI 7 and 14. This dip was significant at DPI 7 for infections of
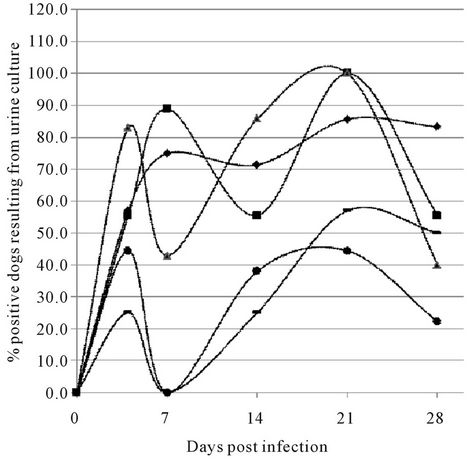
Figure 1. Dynamics of culturing from urine samples of experimentally infected dogs. (Results of urine culture post experimental infection with L. interrogans serovars Copenhageni and Canicola and L. kirschneri SvC Bananal-Liangguang. Number of positive cultures compared to the total are indicated in % for each group of dogs and each serovar as follows: Young dogs infected with Canicola (Y.dogCa) (♦), adult dogs infected with Canicola (A.dogCa) (■), young dogs infected with Copenhageni (Y.dogCo) (▲), adult dogs infected with Copenhageni (A.dogCo) (●) and young dogs infected with SvC Bananal-Liangguang (Y.dogCBL) (▬). Mean values (in %) and spread (95% confidence intervals) are as follows: Y.dogCa. DPI3 = 57 (25 - 84), DPI7 = 75 (41 - 93), DPI14 = 71 (36 - 92), DPI21 = 86 (49 - 97), DPI28 = 83 (44 - 97); A.dogCan. DPI3 = 56 (26.7 - 81.1), DPI7 = 89 (56.5 - 98), DPI4 = 56 (26.7 - 81.1), DPI21 = 100 (70 - 100), DPI28 = 56 (26.7 - 81.1); Y.dogsCo. DPI3 = 83 (34.4 - 87), DPI7 = 43 (15.8 - 75), DPI14 = 86 (48.7 - 97.4), DPI21 = 100 (64.6 - 100), DPI28 = 40 (12 - 77); A. dogs Co. DPI3 = 44 (18.9 - 73.3), DPI7 = 0, DPI14 = 38 (14 - 69), DPI21 = 44 (18.9 - 73.3), DPI28 = 22 (6.3 - 54.7); Y.dogCBL DPI3 = 25 (7.1 - 59.1), DPI7 = 0, DPI14 = 25 (7.1 - 59.1), DPI21 = 57 (25 - 84), DPI28 = 50 (21.5 - 78.5)).
adult dogs with Icterohaemorrhagiae (p = 0.041) and presents a trend in the other cases.
We argue that the unavailability of samples collected between these two time points did not allow a more accurate determination of the actual dips in the various curves, which might very well be similar both in magnitude and in time [27]. Adherence of the leptospires in the lumen will lead to immobilization and hence a transient reduction of their shedding with the urine, thus explaining the decreasing number of positive urine samples collected at DPI 28 [31-33].
Fine tuning of the effects of the kidney colonization by the distinct serovars and their eventual intermittent shedding would need a much longer follow up of urine samples, which was beyond the scope of this paper. Instead, for assessment of the colonization culturing from kidney tissue at DPI 28 was performed. Results should be considered with some caution because early patchy colonization affecting the load of leptospires in the inoculums and cell lysis affecting the viability of leptospires may hamper the quantification of colonized kidneys through the number of positive cultured samples.
Even when taking this into account, the percentage of Canicola positive cultures was markedly higher than that of Copenhageni, while Bananal-Liangguang infections yielded 0% positive cultures (Figure 2). This probably reflects the higher adaptive power of Canicola to dogs that act as its maintenance host compared to the other serovars [13]. This is best illustrated by infections with Bananal-Liangguang. While kidney cultures were all negative, almost half of the urine cultures were positive up to DPI 28 (Table 2), suggesting the presence of the leptospires in the urinary tract without adherence to the renal epithelial cells.
The results obtained by culturing from urine samples, clearly confirmed that the experimental infections in both study A and B were successful. In study B, EDTA hampered interpretation of bacteraemia, but yet it can be concluded that experimental infection was valid, since the majority of the dogs had renal infection as shown by urinary shedding from DPI 14 onwards. It was shown that PCR is a valuable adjunct to culturing for following experimental infections and presents a novel useful tool for the diagnosis of leptospirosis in dogs and probably in other susceptible domestic and farm animals.

Figure 2. Culturing from kidney samples of experimentally infected dogs (results of culturing kidney samples collected 28 days post infection from young and adult dogs infected with L. interrogans serovars Copenhageni, strain CF1 (Y.dogCo and A.dogCo, respectively) and Canicola, strain Moulton (Y.dogCa and A.dogCa, respectively) and young dogs infected with L. kirschneri serovar complex Bananal-Liangguang, strain 11808 (Y.dogCBL)).
4. Acknowledgements
We would like to thank Dr. Mirjam I. Bakker, Epidemiology section, KIT Biomedical Research, for her help with statistical analysis.
REFERENCES
- W. A. Ellis, “Control of Canine Leptospirosis in Europe: Time for a Change?” Veterinary Record, Vol. 167, No. 16, 2010, pp. 602-605.doi:10.1136/vr.c4965
- S. Faine, B. Adler, S. P. Gale, et al., “Leptospira and Leptospirosis,” MediSci, Melbourne, 1999.
- C. Fischer-Tenhagen, C. Hamblin, S. Quandt, et al., “Serosurvey for Selected Infectious Disease Agents in FreeRanging Black and White Rhinoceros in Africa,” Journal of Wildlife Diseases, Vol. 36, No. 2, 2000, pp. 316-323.
- P. N. Levett, “Leptospirosis,” Clinical Microbiology Reviews, Vol. 14, No. 2, 2001, pp. 296-326. doi:10.1128/CMR.14.2.296-326.2001
- World Organization for Animal Health (OIE), “OIE Manual of Diagnostic Tests and Vaccines for Terrestrial Animals,” 15th Edition, Office International des Epizooties, 2004.
- G. M. Cerqueira and M. Picardeau, “A Century of Leptospira Strain Typing,” Infection, Genetics and Evolution, Vol. 9, No. 5, 2009, pp. 760-768. doi:10.1016/j.meegid.2009.06.009
- B. Adler and M. A. de la Pena, “Leptospira and Leptospirosis,” Veterinary Microbiology, Vol. 140, No. 3-4, 2010, pp. 287-296. doi:10.1016/j.vetmic.2009.03.012
- R. Bharadwaj, A. M. Bal, S. A. Joshi, et al., “An Urban Outbreak of Leptospirosis in Mumbai, India,” Japanese Journal of Infectious Diseases, Vol. 55, No. 6, 2002, pp. 194-196.
- A. R. Bharti, J. E. Nally, J. N. Ricaldi, et al., “Leptospirosis: A Zoonotic Disease of Global Importance,” The Lancet Infectious Diseases, Vol. 3, No. 12, 2003, pp. 757- 771.doi:10.1016/S1473-3099(03)00830-2
- S. B. Agampodi, S. J. Peacock, V. Thevanesam, et al., “Leptospirosis Outbreak in Sri Lanka in 2008: Lessons for Assessing the Global Burden of Disease,” The American Journal of Tropical Medicine and Hygiene, Vol. 85, No. 3, 2011, pp. 471-478. doi:10.4269/ajtmh.2011.11-0276
- A. M. Monahan, I. S. Miller and J. E. Nally, “Leptospirosis: Risks during Recreational Activities,” Journal of Applied Microbiology, Vol. 107, No. 3, 2009, pp. 707-716. doi:10.1111/j.1365-2672.2009.04220.x
- B. Abela-Ridder, R. Sikkema and R. A. Hartskeerl, “Estimating the Burden of human Leptospirosis,” International Journal of Antimicrobial Agents, Vol. 36, Suppl. 1, 2010, pp. S5-S7. doi:10.1016/j.ijantimicag.2010.06.012
- E. G. Hartman, P. Franken, B. A. Bokhout, et al., “Leptospirosis in Cattle; Milker’s Fever in Cattle Farmers,” Tijdschrift Voor Diergeneeskunde, Vol. 114, No. 3, 1989, pp. 131-135.
- J. F. Prescott, D. Key and M. Osuch, “Leptospirosis in Dogs,” The Canadian Veterinary Journal, Vol. 40, No. 6, 1999. pp. 430-431.
- M. Ribotta, M. Fortin, R. Higgins, et al., “Canine Leptospirosis: Serology,” The Canadian Veterinary Journal, Vol. 41, No. 6, 2000, pp. 494-495.
- J. E. Sykes, K. Hartmann, K. F. Lunn, et al., “2010 ACVIM Small Animal Consensus Statement on Leptospirosis: Diagnosis, Epidemiology, Treatment, and Prevention,” Journal of Veterinary Internal Medicine, Vol. 25, No. 1, 2011, pp. 1-13. doi:10.1111/j.1939-1676.2010.0654.x
- S. Rahelinirina, A. Leon, R. A. Hartskeerl, et al., “First Isolation and Direct Evidence for the Existence of Large Small-Mammal Reservoirs of Leptospira sp. in Madagascar,” PLoS One, Vol. 5, No. 11, 2010, p. e14111. doi:10.1371/journal.pone.0014111
- F. Merien, P. Amouriaux, P. Perolat, et al., “Polymerase Chain Reaction for Detection of Leptospira spp. in Clinical Samples,” Journal of Clinical Microbiology, Vol. 30, No. 9, 1992, pp. 2219-2224.
- A. M. Bal, “Unusual Clinical Manifestations of Leptospirosis,” Journal of Postgraduate Medicine, Vol. 51, No. 3, 2005, pp. 179-183.
- R. U. Palaniappan, Y. F. Chang, C. F. Chang, et al., “Evaluation of Lig-Based Conventional and Real Time PCR for the Detection of Pathogenic Leptospires,” Molecular and Cellular Probes, Vol. 19, No. 2, 2005, pp. 111-117. doi:10.1016/j.mcp.2004.10.002
- A. Ahmed, M. F. Engelberts, K. R. Boer, et al., “Development and Validation of a Real-Time PCR for Detection of Pathogenic Leptospira Species in Clinical Materials,” PLoS One, Vol. 4, No. 9, 2009, p. e7093. doi:10.1371/journal.pone.0007093
- R. A. Stoddard, J. E. Gee, P. P. Wilkins, et al., “Detection of Pathogenic Leptospira spp. through TaqMan Polymerase Chain Reaction Targeting the LipL32 Gene,” Diagnostic Microbiology and Infectious Disease, Vol. 64, No. 3, 2009, pp. 247-255. doi:10.1016/j.diagmicrobio.2009.03.014
- J. Thaipadungpanit, W. Chierakul, V. Wuthiekanun, et al., “Diagnostic Accuracy of Real-Time PCR Assays Targeting 16S rRNA and lipL32 Genes for Human Leptospirosis in Thailand: A Case-Control Study,” PLoS One, Vol. 6, No. 1, 2011, p. e16236. doi:10.1371/journal.pone.0016236
- B. Victoria, A. Ahmed, R. L. Zuerner, et al., “Conservation of the S10-spc-α Locus within Otherwise Highly Plastic Genomes Provides Phylogenetic Insight into the Genus Leptospira,” PLoS One, Vol. 3, No. 7, 2008, p. e2752. doi:10.1371/journal.pone.0002752
- H. L. Klaasen, M. J. Molkenboer, M. P. Vrijenhoek, et al., “Duration of Immunity in Dogs Vaccinated against Leptospirosis with a Bivalent Inactivated Vaccine,” Veterinary Microbiology, Vol. 95, No. 1-2, 2003, pp. 121-132. doi:10.1016/S0378-1135(03)00152-4
- S. Subharat, P. R. Wilson, C. Heuer, et al., “Evaluation of a SYTO9 Real-Time Polymerase Chain Reaction Assay to Detect and Identify Pathogenic Leptospira Species in Kidney Tissue and Urine of New Zealand Farmed Deer,” Journal of Veterinary Diagnostic Investigation, Vol. 23, No. 4, 2001, pp. 743-752.
- E. S. Broughton and J. Scarnell, “Prevention of Renal Carriage of Leptospirosis in Dogs by Vaccination,” Veterinary Record, Vol. 117, No. 12, 1985, pp. 307-311. doi:10.1136/vr.117.12.307
- P. Schreiber, V. Martin, W. Najbar, et al., “Prevention of Renal Infection and Urinary Shedding in Dogs by a Leptospira Vaccination,” Veterinary Microbiology, Vol. 108, No. 1-2, 2005, pp. 113-118. doi:10.1016/j.vetmic.2005.03.007
- R. M. Nervig and H. C. Ellinghausen Jr., “Viability of Leptospira interrogans Serotype Grippotyphosa in Swine Urine and Blood,” Cornell Veterinary, Vol. 68, No. 1, 1978, pp. 70-77.
- J. M. Minke, R. Bey, J. P. Tronel, et al., “Onset and Duration of Protective Immunity against Clinical Disease and Renal Carriage in Dogs Provided by a Bi-Valent Inactivated Leptospirosis Vaccine,” Veterinary Microbiology, Vol. 137, No. 1-2, 2009, pp. 137-145. doi:10.1016/j.vetmic.2008.12.021
- W. I. Morrison and N. G. Wright, “Canine Leptospirosis: An Immunopathological Study of Interstitial Nephritis Due to Leptospira Canicola,” The Journal of Pathology, Vol. 120, No. 2, 1976, pp. 83-89. doi:10.1002/path.1711200204
- N. F. Cheville, R. Huhn and R. C. Cutlip, “Ultrastructure of Renal Lesions in Pigs with Acute Leptospirosis Caused by Leptospira Pomona,” Veterinary Pathology, Vol. 17, No. 3, 1980, pp. 338-351.
- V. Sitprija, V. Pipatanagul, K. Mertowidjojo, et al., “Pathogenesis of Renal Disease in Leptospirosis: Clinical and Experimental Studies,” Kidney International, Vol. 17, No. 6, 1980, pp. 827-836. doi:10.1038/ki.1980.95
- M. G. Goris, J. F. Wagenaar, R. A. Hartskeerl, et al., “Potent Innate Immune Response to Pathogenic Leptospira in Human Whole Blood,” PLoS One, Vol. 6, No. 3, 2011, p. e18279. doi:10.1371/journal.pone.0018279
- S. Faine, “Leptospirosis,” In: L. Collier, A. Balows and M. Sussman, Eds., Topley and Wilson’s Microbiology and Microbial Infections, Arnold, London, 1998, pp. 854-856.
NOTES
*Corresponding author.

