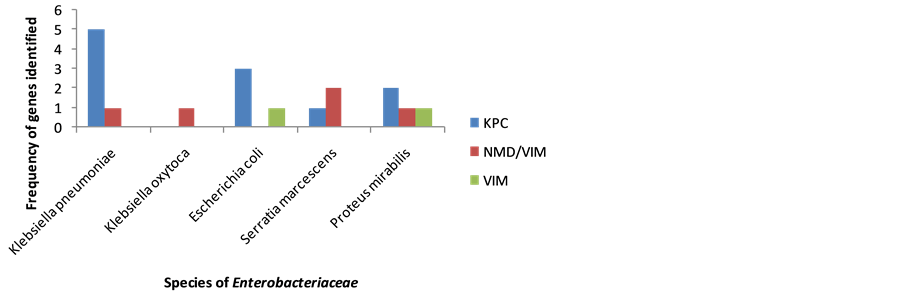Journal of Biosciences and Medicines
Vol.03 No.11(2015), Article ID:61338,8 pages
10.4236/jbm.2015.311013
Characterization of KPC, NDM and VIM Type Carbapenem Resistance Enterobacteriaceae from North Eastern, Nigeria
Yahaya Mohammed1, Sambo B. Zailani2, Anthony O. Onipede3
1Department of Medical Microbiology, Faculty of Basic Medical Sciences, College of Health Sciences, Usmanu Danfodio University, Sokoto, Nigeria
2Department of Medical Microbiology, College of Medical Sciences, University of Maiduguri, Maiduguri, Nigeria
3Department of Medical Microbiology, Faculty of Basic Medical Sciences, College of Health Sciences, Obafemi Awolowo University, Ile-Ife, Nigeria

Copyright © 2015 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).


Received 21 October 2015; accepted 20 November 2015; published 23 November 2015

ABSTRACT
Introduction and Aim: Carbapenem resistance among species of Enterobacteriaceae has emerged as a global public health problem that adds to the high cost of care, severity and high mortality of otherwise straightforward infections. Governments around the world are devoting efforts to com- bat this important threat. The present study was undertaken in our setting to detect and characterize carbapenem resistance among Enterobacteriaceae. Methodology: Confirmed species of Enterobacteriaceae isolated from 225 patients that were admitted in various units of University of Maiduguri Teaching Hospital (UMTH) Maiduguri were screened for carbapenem resistance with meropenem and ertapenem disc (10 µg, Oxoid, England) using clinical and laboratory standards institute (CLSI) breakpoints. Suspected carbapenemase producers were subjected to confirmation using Modified Hodge Test method. Detection of the carbapenemase genes was done by multiplex PCR using KPC, NDM-1 and VIM primers. Results: A total of 225 clinical isolates of Enterobacteriaceae comprising 73 (32.4%) of Klebsiella pneumoniae, 61 (27.1%) of Escherichia coli, 21 (9.3%) of Proteus mirabilis, 18 (8.0%) of Klebsiella oxytoca, 13 (5.8%) of Morganella morganii, 12 (5.3%) of Citrobacter freundii, 12 (5.3%) of Serratia marcescens, 7 (3.1) of Enterobacter aerogenes, 3 (1.4%) of Klebsiella ozaenae, 3 (1.4%) of Hafnia alvei and 2 (0.9%) of Citrobcter sedlakii were isolated. A total of 28 (12.4%) of the isolates screened positive as carbapenemase producers. All the 28 screened isolates were further subjected to confirmation using the Modified Hodge Test for which 23 (10.2%) were confirmed resistant. Therefore a prevalence of 10.2% for carbapenem resistance was recorded in this study. Based on multiplex polymerase chain reaction, the various percentage genotypes of the carbapenemase producers were: 11 (47.8%) for KPC, 2 (8.7%) for VIM while 5 (21.7%) isolates have co-existence of the NDM-1 and VIM genes. However, 5 (21.7%) of the isolates have none of the genes screened for in them. The occurrence of carbapenemase genes among species of Enterobacteriaceae was as follows: 6 (33.3%) for Klebsiella pneumoniae, 4 (22.2%) for Escherichia coli, 4 (22.2%) for Proteus mirabilis, 3 (16.7%) for Serratia marcescens and then 1 (5.6%) for Klebsiella oxytoca. Conclusion: Despite the low use of carbapenem agents in the study area, carbapenem resistance was documented. This calls for an ongoing surveillance and infection control practices of this and other emerging resistance threat in all health centers of Nigeria. Irrational use of antibiotics must be discouraged so as to reduce this resistance threat. Antibiotic stewardship program should be established in all tertiary health centers of Nigeria.
Keywords:
Imipenem, Detection, Genes, Meropenem, Antibiotic

1. Introduction
Gram negative bacteria that produce the extended spectrum beta lactamase enzymes capable of hydrolysing most cephalosporins have been reported worldwide [1] . The carbapenems, namely: imipenem, meropenem, ertapenem and doripenem became the antimicrobials of last resort used in treating infections due to these highly drug resistant bacteria [2] . These antimicrobial agents became crucial in the management of life threatening healthcare associated and community acquired infections. The consequence of this widespread use of carbapenems was the emergence of the first carbapenemase producing Enterobacteriaceae (CRE) in 1993 [1] .
The major risk factors for acquiring these CRE were: organ or stem cell transplantation, intensive care unit admission, poor nutritional status, severe illness, mechanical ventilation, prolonged hospitalization and previous surgery [3] . The centre for disease control and prevention (CDC) recommends rapid action for CRE in terms of proper/timely identification and the institution of aggressive preventive measures to combat this emerging threat [4] . The present study was undertaken to detect and characterize CRE from a major reference centre in north- eastern Nigeria using both the phenotypic and genotypic methods.
2. Methodology
2.1. Study Area
University of Maiduguri Teaching Hospital (UMTH), Borno State. The principal referral center of northeastern Nigeria.
2.2. Study Design
The study was a hospital based, descriptive and cross-sectional.
2.3. Study Population
The study was carried out on patients hospitalized in UMTH from the following wards; medical ward (male and female), surgical ward (male and female), special care baby unit (SCBU) and the intensive care unit (ICU).
2.4. Study Period
The study was carried out from June 2014 to December 2014.
2.5. Sampling Method
Convenient (non-probability type) sampling.
2.6. Specimen Collection
The isolates of Enterobacteriaceae were obtained from the following specimens; blood, urine, cerebrospinal fluid, stool, and swabs of patients with invasive diseases (i.e. bloodstream infections, catheter related infections, ventilator assisted infections, etc.) All specimen were collected and transported according to standard methods [5] .
2.7. Bacterial Identification
The specimen were inoculated on MacConkey agar. After 24 - 48 hours of aerobic incubation at 36˚C - 37˚C, colonial appearance and characteristics of isolates on MacConkey agar was noted and they were then subjected to Gram staining reaction and also motility testing according to standard methods [5] . All suspected isolates of Enterobacteriaceae were confirmed by the Microbact Gram negative identification system™ (Oxoid) 24E according to the manufactures instructions.
2.8. CRE Screening and Phenotypic Confirmation
All the bacterial isolates were screened for carbapenemases according to the CLSI guidelines [6] . Ertapenem and meropenem discs (10 μg, Oxoid, England) were used. The antibiotic discs were placed on the surface of inoculated Mueller Hinton Agar plates using a sterile forceps. The discs were placed about 30 mm apart and the plates were incubated for 24 hours at 37˚C after which zones of inhibitions were read.
Isolates that showed a zone of inhibition ≤ 21 mm in diameter for meropenem and/or ertapenem were considered as suspected carbapenemase producers. Escherichia coli ATCC 25922 was used to quality control the screening testing [6] .
All Enterobacteriaceae isolates which were found resistant to Meropenem (10 μg) disc and or Ertapenem (10 μg) disc, either alone or in totality were subjected to confirmatory testing using the Modified Hodge Test [6] .
A 0.5 McFarland standard of Escherichia coli ATCC® 25922 was inoculated in saline and diluted to 1:10 in saline. This suspension was then evenly inoculated with a sterile cotton swab on surface of Mueller Hinton Agar plate. A disc of Ertapenem (10 μg, Oxoid, England) was placed on the surface of Mueller Hinton agar plates at the centre. Thereafter, by means of a sterilized wire loop, the test organism was streaked together with the two quality control organisms (Klebsiella pneumoniae ATCC® BAA-1705―MHT positive and Klebsiella pneumoniae ATCC® BAA-1706―MHT negative) in a straight line out from the edge of the Ertapenem disc. The plates were then incubated at 37˚C for 24 hours. Positivity for carbapenem was inferred when there is the appearance of a clover leaf type indentation or flattening at the intersection of the test organism and Escherichia coli ATCC 25922 within the zone of inhibition of the carbapenem susceptibility disc as described by Anderson and recommended by CLSI [6] [7] . Klebsiella pneumoniae ATCC® BAA-1705-MHT positive and Klebsiella pneumoniae ATCC® BAA-1706-MHT negative were used as positive and negative controls for MHT respectively [6] .
2.9. DNA Extraction
The Quick Extract™ bacterial DNA extraction kit was used to extract DNA from bacterial cells using a solid phase enzymatic method. A loopful of the organism was harvested from fresh colony of the bacterial species grown on MacConkey agar. DNA extraction was subsequently done according to manufacturer’s instructions [8] . Extracted DNA was then eluted from the columns in 100 μL of elution buffer and stored at −20˚C for further PCR analysis.
2.10. Primer Sequence
PCR analysis for beta lactamase genes of the family KPC, NDM and VIM was carried out. Primers were obtained from Bioneer, Inc. Company, USA. The resistance genes blaKPC, blaNDM and blaVIM were amplified by PCR using previously published primers [9] -[12] .
2.11. DNA Amplification
The prepared PCR products with master mixture were placed in the eppendrof thermal cycler. Amplification was carried out according to standard thermal and cycling condition.
2.12. Data Analysis
Data collected was recorded into a computer and analysis was be done using statistical package for social sciences version 16.0 (SPSS Chicago Ill. USA). Results were presented when necessary as tables, figures, diagrams and photograph.
2.13. Ethical Consideration
The study protocol was reviewed and approved by the ethical review committee of UMTH.
3. Results
A total of 225 isolates were identified as Gram negative rods (Enterobacteriaceae). The distribution of the various species of the 225 Enterobacteriaceae isolated were as follows; 73 (32.4%) for Klebsiella pneumoniae, 61 (27.1%) for Escherichia coli, 21 (9.3%) for Proteus mirabilis, 18 (8.0%) for Klebsiella oxytoca, 13 (5.8%) for Morganella morganii, 12 (5.3%) for Citrobacter freundii, 12 (5.3%) for Serratia marcescens, 7 (3.1%) for Enterobacter aerogenes, 3 (1.4%) for Klebsiella ozaenae, 3 (1.4%) for Hafnia alvei and 2 (0.9%) for Citrobacter sedlakii.
A total of 115 (51.1%) clinical isolates of Enterobacteriaceae were obtained from urine, 45 (20.0%) from blood specimen, 31 (13.8) from swabs, 21 (9.3%) from sputum and 13 (5.8%) from cerebrospinal fluid.
The carbapenem susceptibility status of Enterobacteriaceae following preliminary screening with meropenem and ertapenem were as shown in Table 1.
All the 225 species of Enterobacteriaceae were screened for carbapenem resistance using meropenem and ertapenem disc. 22 (9.7%) of the various species of Enterobacteriaceae were resistant to meropenem while 28 (12.4%) were resistant to ertapenem.
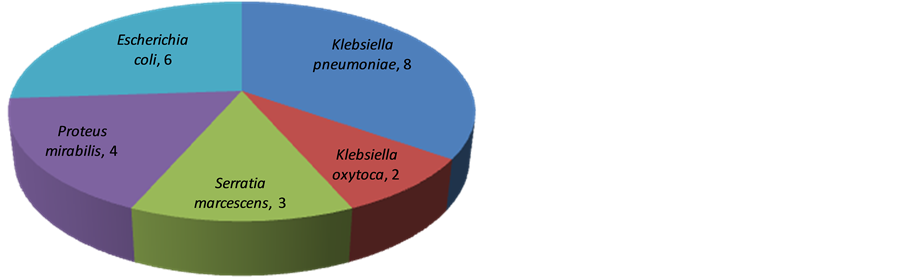
The 28 isolates of Enterobacteriaceae that tested positive to the screening test were subjected to confirmatory test using MHT as shown in Table 2. A total of 23 (82.1%) of the 28 (100%) isolates tested positive using the MHT. Figure 1 shows the distribution of carbapenem resistance among species of Enterobacteriaceae following the Modified Hodge Test.
All the 23 isolates were further characterized for their molecular genotype by the use of PCR. Using DNA primers for KPC, NDM-1 and VIM genes; a total of 11 (47.8%) KPC and 2 (8.8%) VIM genes were detected in 18 (78.3%) of the 23 (100%) isolates. However, 5 (21.7%) of the isolates have NDM-1 and VIM gene coexisting together. No gene was detected in 5 (21.7%) of the isolates.
Table 1. Carbapenem susceptibility status of Enterobacteriaceae following preliminary screening with meropenem and ertapenem (N = 225).
S = Sensitive, R = Resistance and T = Total.
Table 2. Modified hodge test on screened carbapenem resistant Enterobacteriaceae (N = 28).
Figure 1. The distribution of carbapenem resistance in species of Enterobacteriaceae following the modified hodge test (N = 23).
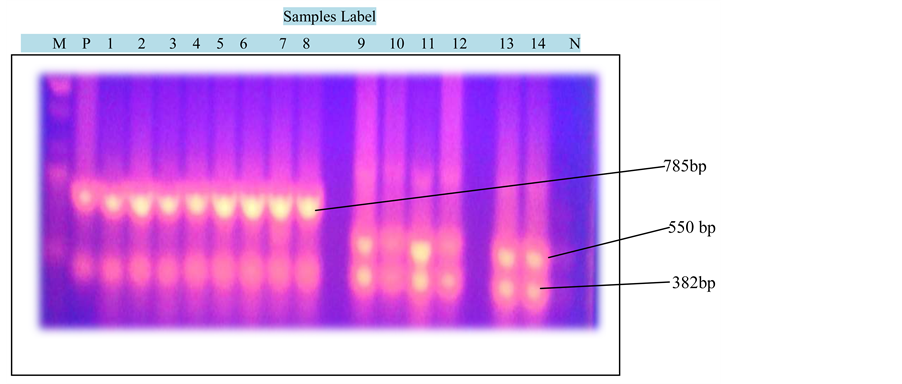
Figure 2 shows the photograph of PCR products of carbapenemase genes. KPC gene was detected at 785 bp, NDM-1 at 550 bp and VIM at 382 bp genes. The DNA ladder was set at 100 bp. The distribution of the carbapenemase genes among various species of Enterobacteriaceae is shown in Figure 3.
The distribution of the 11 (100%) KPC genes within the various organisms was as follows; 5 (45.5%) for Klebsiella pneumoniae, 3 (27.3%) for Escherichia coli, 2 (18.2%) for Proteus mirabilis and 1 (9.0%) for Serratia marcescens. The distribution of the 2 (100%) VIM genes within the various organisms were as follows 1 (50.0%) each for Escherichia coli and Proteus mirabilis. The 5 (100%) genes of NMD-1 and VIM that coexisted together were distributed in 2 (40%) of the Serratia marcescens, 1 (20%) each for Klebsiella pneumoniae, Klebsiella oxytoca and Escherichia coli. No genes were detected in 2 (40%) of the Klebsiella pneumoniae, 1 (20%) of Klebsiella oxytoca and 2 (40%) of the Escherichia coli.
4. Discussion
The occurrence of carbapenems in the north-eastern part of Nigeria was established from this study. Following preliminary screening with meropenem and ertapenem, a resistance of 22 (9.7%) and 28 (12.4%) were reported respectively. This was similar to a prevalence reported by Yusuf et al. in Kano among isolates of Enterobacteriaceae to imipenem and meropenem of 10.5% and 12.5% respectively following phenotypic screening from surgical and intensive care units [13] . The same author reported a 7.4% resistance to imipenem and 87.5% resistance to meropenem among Enterobacteriaceae using the same study design from the same institution but in a different patient population [14] .
This showed that with difference of sample population even within the same centre, a different resistance pattern might be shown as was the case from this study. The epidemiologic significance of the finding for carbapenems resistance following preliminary screening as observed from this study confirms the existence of carbapenems resistance from our locality. Hence, this calls for ongoing surveillance of this resistance threat in our healthcare setting. The confirmatory test (Modified Hodge test) detected 23 (10.2%) as carbapenemase producers out of the 28 screened carbapenem resistant Enterobacteriaceae. This means that a prevalence of 23 (10.2%) was detected from this study. This was slightly lower than a prevalence of 14.0% recorded in Kano by Yusuf et al. among species of Enterobacteriaceae with the highest prevalence found among Klebsiella pneumoniae (16.7%), followed by Proteus species (16.0%), and Escherichia coli (12.5%) while no carbapenemases were detected in Serratia species [14] . A much lower prevalence of carbapenemases of 0.15% was reported by Jones in Israel using the MHT [15] .
The finding of a high prevalence of carbapenemases in Klebsiella pneumoniae from this study agrees with the finding of Landman [16] who reported that over one-third of Klebsiella pneumoniae collected in 2006 in New York, USA carried the carbapenemase enzyme. Several studies in the US have reported carbapenem resistant Klebsiella pneumoniae as the specie most commonly encountered there [17] .
Although the study area; UMTH Maiduguri, Borno State, north-eastern Nigeria does not have a regular prescription pattern for carbapenem probably due to their high costs nevertheless a relative high prevalence of CRE was detected. This was not surprising because numerous studies have shown that prior carbapenem therapy is not a pre-requisite for carbapenem resistance among Enterobacteriaceae [18] .
Figure 2. Photograph of PCR products of carbapenemase genes: KPC (785 bp), NDM-1 (550 bp) and VIM (382 bp) genes. DNA marker levels from 100 bp to 1500 bp and samples 1 to 14 with M (DNA Ladder), P (Positive control) and N (Negative Control).
Figure 3. The distribution of the carbapenemase genes among species of Enterobacteriaceae.
However, it is important to note that substantial amount of patients that attend UMTH Maiduguri are usually being referred from other hospitals in the region, probably this resistant pathogens were imported from other regions of Nigeria or from neighboring countries due to increase in local and international travels.
In this study, the occurrences and molecular features of the carbapenemase genes; KPC, NDM-1 and VIM in the north-eastern part of Nigeria were established. The finding of NDM-1 from this study is a worrisome fact, this is because NDM-1 has the ability to spread, unlike any resistant mechanism that has ever been seen in clinical microbiology [19] . KPC gene was the predominant carbapenemase gene detected in this study. Although, KPC gene was reported to be the predominant carbapenemases among Enterobacteriaceae worldwide, [20] Spellberg and colleagues reported that NDM-1 was predominantly common in survey of Enterobacteriaceae they did in 2009 but in 2010 they reported KPC as the most predominant in UK [21] .
The co-existence of NDM-1 and VIM genes were noted from this study. The clinical significance of this finding is that patients having organisms possessing this double genes are more likely to have multi drug resistance and more likely to have the propensity for widespread nosocomial transmission. In the present study, 5 (21.7%) of the isolates had none of the genes detected in them. This implies that other factors, other than the presence of KPC, NDM-1, VIM were effective in producing resistance to carbapenem group of antibiotics.
5. Conclusions
The present study highlights the existence of carbapenem resistance among Enterobacteriaceae in a center that does not routinely use carbapenem antibiotics. The carbapenemase producing pathogens were simultaneously resistant to meropenem and ertapenem disc using the disc diffusion method.
In view of this emerging drug resistance, the practice of routine CRE testing along with conventional antibiogram would be useful for all cases which will help in the proper treatment of the patient and also prevent further development of bacterial drug resistance. The role of infection control measures and antibiotic stewardship programs cannot be overemphasized.
Cite this paper
YahayaMohammed,Sambo B.Zailani,Anthony O.Onipede, (2015) Characterization of KPC, NDM and VIM Type Carbapenem Resistance Enterobacteriaceae from North Eastern, Nigeria. Journal of Biosciences and Medicines,03,100-107. doi: 10.4236/jbm.2015.311013
References
- 1. Patrice, N., Laurent, P. and Laurent, D. (2012) Rapid Detection of Carbapenemase Producing Enterobacteriaceae. Emerging Infectious Diseases, 18, 1503-1507.
http://dx.doi.org/10.3201/eid1809.120355 - 2. Mammeri, H., Guillon, H.E. and Nordmann, P. (2010) Phenotypic and Biochemical Comparison of the Carbapenems Hydrolyzing Activities of Five Plasmid-Borne AmpC Beta Lactamases. Antimicrobial Agents and Chemotherapy, 54, 4556-4560.
http://dx.doi.org/10.1128/AAC.01762-09 - 3. Olaitan, A.O., Berrazeg, M., Fagade, O.E., Adelowo, O.O., Alli, J.A. and Rolain, J.M. (2013) Emergence of Multi Drug Resistant Acinetobacter baumanii Producing OXA-23 Carbapenemase, Nigeria. International Journal of Infectious Diseases, 17, 469-470.
http://dx.doi.org/10.1016/j.ijid.2012.12.008 - 4. Centre for Disease Control (2013) Stop Infections from Lethal CRE Germs Now. CDC Vital Signs Report.
http://www.cdc.gov/hai/organisms/cre/TrackingCRE.html - 5. Winn, W., Allen, S., Janda, W., Koneman, E., Procop, G., Schreckenberger, P., et al. (2006) Introduction. Koneman’s Color Atlas and Text Book of Diagnostic Microbiology. Lippincott Williams & Wilkins, Baltimore and Philadelphia, 21-25.
- 6. CLSI (2011) Performance Standards for Antimicrobial Susceptibility Testing: Twenty-First Informational Supplement. CLSI Document M100-S21. Clinical and Laboratory Standard Institute, Wayne.
- 7. Anderson, K.F., Lonsway, D.R., Rasheed, J.K., Biddle, J., Jensen, B. and McDougal, L.K. (2007) Evaluation of Methods to Identify the Klebsiella pneumoniae Carbapenemase in Enterobacteriaceae. Journal of Clinical Microbiology, 45, 2723-2725.
http://dx.doi.org/10.1128/JCM.00015-07 - 8. Van-Tongeren, S.P., Degener, J.E. and Harmsen, H.J.M. (2011) Comparison of Three Rapid and Easy Bacterial DNA Extraction Methods for Use with Quantitative Real Time PCR. European Journal of Clinical Microbiology & Infectious Diseases, 30, 1053-1061.
http://dx.doi.org/10.1007/s10096-011-1191-4 - 9. Nordmann, P., Poirel, L., Carrer, A., Toleman, M.A. and Walsh, T.R. (2011) How to Detect NDM-1 Producers. Journal of Clinical Microbiology, 49, 718-721.
http://dx.doi.org/10.1128/JCM.01773-10 - 10. Ellington, M.J., Kistler, J., Livermore, D.M. and Woodford, N. (2007) Multiplex PCR for Rapid Detection of Genes Encoding Acquired Metallo-Beta-Lactamases. Journal of Antimicrobial Chemotherapy, 59, 321-322.
http://dx.doi.org/10.1093/jac/dkl481 - 11. Pillai, D.R., Melano, R., Rawte, P., Lo, S., Tijet, N., Fuksa, M., et al. (2009) Klebsiella pneumoniae Carbapenemase, Canada. Emerging Infectious Diseases, 15, 827-829.
http://dx.doi.org/10.3201/eid1505.081536 - 12. Anna, K.M., Inneke, V.H., Flavia, R., Anna, S.L. and Silvia, F.C. (2009) Multiplex PCR for Rapid Detection of Genes Encoding Oxacillinase and Metallo-Beta-Lactamases in Carbapenem Resistant Acinotobacter Species. JMM Correspondence, 10, 1522-1524.
http://jmm.sgmjournals.org - 13. Yusuf, I., Arzai, A.H., Getso, M.I., Sherif, A. and Haruna, M. (2012) Emergence of Carbapenem Resistant Enterobacteriaceae in Surgical and Intensive Care Units of a Hospital with Low Usage of Carbapenem in Kano, North West Nigeria. Antimicrobial Resistance and Infection Control, 2, 75-78.
http://dx.doi.org/10.1186/2047-2994-2-S1-P75 - 14. Yusuf, I., Magashi, A.M., Firdausi, F.S., Sharif, A.A., Getso, M.I., Bala, J.A., et al. (2012) Phenotypic Detection of Carbapenemases in Members of Enterobacteriacea in Kano, Nigeria. International Journal of Science and Technology, 2, 802-806.
- 15. Jones, R.N., Biedenbach, D.J., Sader, H.S., Fritsche, T.R., Toleman, M.A. and Walsh, T.R. (2005) Emerging Epidemic of Metallo-Beta Lactamases Mediated Resistance. Diagnostic Microbiology and Infectious Diseases, 51, 77-84.
http://dx.doi.org/10.1016/j.diagmicrobio.2004.07.003 - 16. Landman, D., Bratu, S., Kochar, S., Panwar, M., Trehan, M., Doymaz, M. and Quale, J. (2007) Evolution of Antimicrobial Resistance among Pseudomonas aeruginosa, Acinetobacter baumannii and Klebsiella pneumoniae in Brooklyn, NY. Journal of Antimicrobial Chemotherapy, 60, 78-82.
http://dx.doi.org/10.1093/jac/dkm129 - 17. Fernando, P., Tania, M., Leonor, G., Melina, R. and Alejandra, C. (2009) Sensitive Screening Tests for Suspected Class A Carbapenemase Production in Species of Enterobacteriaceae. Journal of Clinical Microbiology, 47, 1631-1639.
http://dx.doi.org/10.1128/JCM.00130-09 - 18. Poirel, L., Benouda, A., Hays, C. and Nordmann, P. (2011) Emergence of NDM-1 Producing Klebsiella pneumoniae in Morocco. Journal of Antimicrobial Chemotherapy, 66, 2781-2783.
http://dx.doi.org/10.1093/jac/dkr384 - 19. Jennifer, C. and Adrian, B. (2011) The Emergence of Carbapenem Resistance in Enterobacteriaceae in South Africa. Southern African Journal of Epidemiology & Infection, 26, 239-240.
- 20. Rapp, R.P. and Urban, C. (2012) Klebsiella pneumoniae Carbapenemases in Enterobacteriaceae: History, Evolution, and Microbiology Concerns. Pharmacotherapy, 32, 399-407.
http://dx.doi.org/10.1002/j.1875-9114.2012.01035.x - 21. Spellberg, B., Blaser, M. and Guidos, R.J. (2011) Combating Antimicrobial Resistance: Policy Recommendations to Save Lives. Clinical Infectious Diseases, 52, 397-405.
http://dx.doi.org/10.1093/cid/cir153