Journal of Cosmetics, Dermatological Sciences and Applications
Vol.05 No.04(2015), Article ID:62307,11 pages
10.4236/jcdsa.2015.54040
Modulation of a Specific Pattern of microRNAs, Including miR-29a, miR-30a and miR-34a, in Cultured Human Skin Fibroblasts, in Response to the Application of a Biofunctional Ingredient that Protects against Cellular Senescence in Vitro
Xianghong Yan1, Catherine Serre2, Laurine Bergeron2, Ludivine Mur2, Valère Busuttil2, Jean-Marie Botto2*, Nouha Domloge2
1P&G Innovation Godo Kaisha, Kobe, Japan
2Ashland Specialty Ingredients, Vincience Global Skin Research Center, Sophia Antipolis, France

Copyright © 2015 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 1 December 2015; accepted 25 December 2015; published 29 December 2015
ABSTRACT
Skin aging is a process of structural and compositional remodeling that can be manifested by wrinkling and sagging. Remarkably, the dermis plays a dominant role in the aging process. Recent studies suggest that microRNAs are implicated in the regulation of gene expression during aging. However, studies about age-related microRNAs and how they modulate skin aging remain limited. In the present work, a complex of hydrolyzed natural yeast proteins (Saccharomyces cerevisiae) and hydrolyzed natural soya bean was developed and showed the ability to modulate the expression of telomere-binding protein TRF2, which is a key factor for telomere protection and to prevent cellular senescence in vitro and DNA damage. The aim of the study was to identify microRNAs specifically modulated after application of the ingredient complex to cultured fibroblasts, and their possible involvement in remodeling of the human extracellular matrix and fibroblast senescence. Consequently, human skin fibroblasts were cultured and treated with 1% of the ingredient complex for 48 h before analyzing microRNA modulation by RT-qPCR. The use of bioinformatics allowed us to predict the target genes for modulated microRNAs. Results show that the ingredient complex modulated a pattern of microRNAs including the down-regulation of miR-29a-3p, miR-30a-5p and miR-34a-5p, which are associated with fibroblast senescence and remodeling of the human dermal extracellular matrix. In conclusion, our results indicate that miR-29a-3p, miR-30a-5p and miR-34a-5p possibly represent key microRNAs that impact human fibroblast senescence and remodeling of the dermal extracellular matrix.
Keywords:
microRNAs, qPCR Array, Skin Fibroblasts, Skin Senescence, Telomere, Extracellular Matrix, Sirtuin 1, Collagen

1. Introduction
The skin is the largest organ of the human body and fulfills many important functions. It plays a vital role in protecting the internal organs from external aggression and environmental factors such as trauma, ultraviolet and infrared radiation, heat, environmental pollution and microbial pathogens. As a consequence of the increase in life expectancy, healthy aging is a matter of concern. At the level of the skin, the consequences of aging can lead to a weaker barrier with reduced protection favoring disease conditions that affect quality of life as well as physical appearance. Hence, modulating the progression of skin aging has become increasingly important.
From a physiological point of view, skin aging is a structural and compositional remodeling associated with phenotypic changes in skin cells. Aged skin has a thinner epidermis and a decrease in both the quantity and quality of the dermal extracellular matrix (ECM). Remarkably, the dermis plays a dominant role in the process of skin aging. While this process is subject to both intrinsic and extrinsic factors, the consequences involve abnormal synthesis of proteins in the dermal extracellular matrix, increased degradation of collagen and elastin, and senescence of dermal fibroblasts.
Recent studies suggest that microRNAs (miRs) also play a role in the regulation of gene expression in organism aging [1] [2] . MicroRNAs are a class of short, endogenous, single-stranded, non-coding RNA molecules [3] . By binding to specific 3’-UTR sequences of target mRNAs, microRNAs can regulate the expression of multiple genes at the post-transcriptional level through degradation or translational inhibition of the targeted transcripts [4] . Some recent studies have compared young and aged organs or tissue such as the heart [5] , cornea [6] and kidneys [7] , and pointed out the involvement of senescence-associated microRNAs in the regulation of the aging process.
In this study, the effect of a specific complex of hydrolyzed natural yeast proteins from Saccharomyces cerevisiae and hydrolyzed natural soya proteins was investigated after its application to cultured skin fibroblasts. Soybean (US) or soya bean (UK) (Glycine max) is a legume species native to East Asia. This annual plant has been used in China for 5000 years as food and a component of medical preparations. Soya is a perfect source of protein with significant amounts of all the essential amino acids for humans, and it is rich in antioxidant isoflavones. Soybean hydrolysate shows chelating and radical scavenging activity, and it decreases lipid peroxidation [8] . Saccharomyces cerevisiae (commonly known as baker’s or brewer’s yeast) is a single-celled eukaryote capable of metabolic respiration and fermentation, making it rich in essential amino acids and small peptides.
This ingredient complex was developed for its ability to positively modulate the expression of TRF2 and protect against UV-induced DNA damage in vitro [9] - [11] . Moreover, this complex was demonstrated to prevent the signs of cellular senescence in vitro, as it was able to prevent the increase of senescence-associated beta- galactosidase induced by a FOXO3a-specific siRNA in cultured human fibroblasts [12] .
Results show that the ingredient complex induced a specific pattern of microRNA down-regulation in vitro. Bioinformatic prediction pointed out potential target genes for the identified microRNAs, particularly SIRT1 and collagens, with a relationship to the modulation of human fibroblast senescence and remodeling of the extracellular matrix. Remarkably, three of the down-regulated microRNAs (miR-29a-3p, miR-30a-5p and miR- 34a-5p) are well described for their implication in cellular senescence and are predicted to target SIRT1 expression.
2. Materials and Methods
2.1. Cell Culture
Normal human neonatal dermal fibroblasts (PromoCell GmbH, Heidelberg, Germany) were cultured in Dulbecco’s modified Eagle’s Medium (DMEM), 1 g/L glucose (Lonza, Rockland, ME, USA) supplemented with 10% decomplemented fetal calf serum (Lonza), 2 mM of L-glutamine (Lonza) and 0.1 mg・ml−1 Primocin™ (Invivogen, San Diego, CA, USA). Fibroblasts were cultured at 37˚C in a humidified atmosphere containing 5% CO2. The tested ingredient (Ashland Specialty Ingredients, Wilmington, USA) was used in our experiments at the dose of 1% (1:100 dilution v:v).
2.2. Properties of the Tested Natural Ingredient Complex
The tested ingredient is a complex of hydrolyzed natural yeast proteins (Saccharomyces cerevisiae) and hydrolyzed natural soya proteins with a total dry matter weight of 7 g/kg. After screening several botanical and yeast extracts and their combination, this ingredient complex was selected for its ability to modulate the expression of telomere-binding proteins such as TRF2 [9] - [11] in ex vivo human skin, human primary keratinocytes and human neonatal fibroblasts in culture. This ingredient is described in patent FR12 00814.
2.3. microRNAs of Interest
The list of microRNAs with a described implication in human dermal homeostasis, fibroblast senescence and regulation of dermal extracellular matrix content was established according to the literature (Table 1). Twenty- one microRNAs were hence selected by compiling the microRNAs described in the literature: miR-10b-5p, miR-1185-5p, miR-1204, miR-143-3p, miR-145-5p, miR-146a-5p, miR-155-5p, miR-199a-3p, miR-21-5p, miR-23a-3p, miR-26a-5p, miR-29a-3p, miR-30a-5p, miR-335-3p, miR-34a-5p, miR-519c-3p, miR-542-5p, miR-638, miR-658, miR-663 and miR-7-5p.
2.4. Target Genes
Bioinformatics was used to obtain a list of predicted and experimentally verified target genes for the selected microRNAs in this study. Two types of analyses were conducted.
i) The first approach used miRWalk, a comprehensive database providing the largest available collection of predicted and experimentally verified (validated) microRNA-target interactions [13] . Each microRNA that was modulated in our experiments was searched in the “validated targets module” of miRWalk in order to retrieve a list of validated targets that was further processed to retain only the genes involved in extracellular matrix homeostasis and fibroblast senescence.
ii) The second approach used Ingenuity® Pathway Analysis (IPA) (Qiagen, Redwood City, CA, USA) to possibly identify new interactions that could be complementary to the ones identified through the miRWalk database. IPA predicts microRNA interactions with target genes, based on the aggregation of data from TarBase (experimentally validated data), miRecords, and TargetScan (predicted microRNA-mRNA interactions) [14] - [16] . The resulting lists of genes established by these two approaches are presented in Table 2.
Table 1. List of studied microRNAs. MicroRNAs implicated in fibroblast senescence were selected according to scientific literature. The described up-regulation (up) or down-regulation (down) during fibroblast senescence is indicated.
Table 2. Relative expression of microRNAs and potential target genes. The relative quantification (RQ) was experimentally measured for each of the 18 microRNAs (column 1) in fibroblasts treated with the ingredient versus untreated fibroblasts (column 2). Four microRNAs (miR-146a-5p, miR-155-5p, miR-21-5p, miR-542-5p) showed RQ values higher than 0.85 and were not considered in the study (microRNAs indicated in brackets). RQ values lower than or equal to 0.85 are highlighted in bold, and correspond to 14 microRNAs (miR-10b-5p, miR-1185-5p, miR-143-3p, miR-145-5p, miR-199a-3p, miR-23a-3p, miR-26a-5p, miR-29a-3p, miR-30a-5p, miR-335-3p, miR-34a-5p, miR-638, miR-663, miR-7-5p). Target genes identified through miRWalk and selected for their potential implication in extracellular matrix homeostasis and fibroblast senescence are indicated in column 3. miR-1185-5p, miR-23a-3p and miR-335-3p showed no validated target genes in miRWalk, reducing to 11 the number of microRNAs of interest (miR-10b-5p, miR-143-3p, miR-145-5p, miR-199a-3p, miR-26a-5p, miR-29a-3p, miR-30a-5p, miR-34a-5p, miR-638, miR-663, miR-7-5p) (column 1, underlined). Ingenuity® Pathway Analysis was also used to complementarily identify potential target genes (column 5). The target genes identified in common by miRWalk and Ingenuity® Pathway Analysis are listed in column 4.
2.5. qPCR Array Experiments
Cells were grown in 100-mm culture dishes and treated or not with 1% tested ingredient in the culture medium for 48 h. The culture medium was then removed and the cells were washed with cold PBS. Total RNA including microRNAs was extracted from each sample using the mirVana miRNA Isolation Kit (Ambion, Austin, TX, USA) and collected in 100 µl of RNase-free water. RNA quantity, quality and purity were determined using a NanoDrop™ 2000 spectrophotometer (Thermo Scientific, Waltham, MA, USA) by measuring the ratio of absorbance at 260 and 280 nm (A260/A280) and 260 and 230 nm (A260/A230). For each sample (untreated and treated), 10 ng of total RNA were reverse-transcribed by using the Universal cDNA Synthesis Kit and miRCURY LNA™ (Exiqon, Vedbaek, Denmark) on an MJ MiniCycler™ thermal cycler (MJ Research, Waltham, MA, USA). Real-time qPCR to amplify the 21 selected microRNAs was performed on a StepOnePlus™ thermal cycler (Applied Biosystems, Foster City, CA, USA) using the Pick-&-Mix microRNA PCR Panel with custom selection of LNA™ microRNA primer sets (Exiqon) and ExiLENT SYBR® Green Master Mix (Exiqon), according to the manufacturer’s recommendations.
The references of the microRNA LNA™ PCR primer sets (Exiqon) used in this study are: miR-10b-5p (ref. 205637), miR-1185-5p (ref. 205926), miR-1204 (ref. 204123), miR-143-3p (ref. 205992), miR-145-5p (ref. 204483), miR-146a-5p (ref. 204688), miR-155-5p (ref. 204308), miR-199a-3p (ref. 204536), miR-21-5p (ref. 204230), miR-23a-3p (ref. 204772), miR-26a-5p (ref. 204772), miR-29a-3p (ref. 204698), miR-30a-5p (ref. 205695), miR-335-3p (ref. 205613), miR-34a-5p (ref. 205613), miR-519c-3p (ref. 204016), miR-542-5p (ref. 204198), miR-638 (ref. 204234), miR-658 (ref. 204234), miR-663 (ref. 205877), and miR-7-5p (ref. 205727). Let-7a-5p (ref. 205727) was used as the reference for analysis. The comparative Ct method was used for relative quantification of microRNA expression.
2.6. Interaction Network of microRNAs and Genes
An interaction network was built using Ingenuity® Pathway Analysis (Qiagen). This network included the 18 microRNAs that showed a modulation in expression after treatment with the ingredient, and the 25 target genes described in the miRWalk database (Table 1). Around these microRNAs and genes, an interaction network was developed: the network components were grouped according to their biological activity (e.g., collagen genes, metalloproteinases, transcription factors such as SOXs, FOXO3 and sirtuins, which are related to senescence regulation, BIRK5 and AURKB components of the chromosomal passenger complex). MicroRNAs were placed at the periphery of the interaction network and predicted relationships with protein-coding genes were established. New relationships (in blue) could be established by the introduction of 8 new components in the network (Figure 2(b), in yellow): nidogen 1 (NID1), integrin beta 6 (ITGB6), integrin beta 8 (ITGB8), collagen IV alpha 5 (COL4A5), ADAM metallopeptidase with thrombospondin type 1 motif, 6 (ADAMTS6), reversion-inducing cysteine-rich protein with Kazal motifs (RECK), chymase 1 (CMA1) and fibroblast growth factor 13 (FGF13).
3. Results
The purpose of the present study was to identify microRNAs involved in fibroblast senescence and modulation of the dermal extracellular matrix structure, and to identify potentially modulated target genes by bioinformatic prediction. A list of microRNAs of interest was obtained following an overview of scientific literature and database searches. Modulation of the expression level of the microRNAs on this list was assessed by microRNA qPCR array, in cultured normal human fibroblast treated or not with the natural ingredient complex.
3.1. Selection of the microRNAs of Interest
Twenty-one microRNAs of interest and relevant to fibroblast physiology were selected according to the literature (Table 1). Among the selected microRNAs involved in fibroblast senescence, some have been described with decreased expression levels during senescence: miR-143-3p, miR-145-5p, miR-155-5p, miR-21- 5p, miR-23a-3p [17] and miR-7-5p [18] . Other microRNAs are known to have increased expression levels during the senescence process: miR-10b-5p, miR-146a-5p, miR-199a-3p, miR-26a-5p, miR-34a-5p, miR-542-5p [17] , miR-1185-5p, miR-1204, miR-519c-3p, miR-658 [18] , miR-29a-3p, miR-30a-5p [19] , and miR-638, miR- 663 [20] . MiR-29-3p is also involved in fibroblast extracellular matrix synthesis and senescence [21] . MiR-335-3p was described as having a role in proliferation, migration and differentiation [22] . These microRNAs are listed in Table 1.
3.2. microRNA qPCR Array
A microRNA qPCR array including the 21 microRNAs of interest was used to test the modulation of these microRNAs after application of the ingredient to cultured normal human fibroblasts for 48 h. Three microRNAs (miR-1204, miR-519c-3p, miR-658) were not detected by qPCR and were thus considered as non-expressed by fibroblasts under our experimental conditions, reducing the list of microRNAs from 21 to 18. Amplification of these 18 microRNAs (Table 2, column 1) showed RQ values between 0.573 (for miR-335-3p) and 0.919 (for miR-155-5p) (Table 2, column 2). Out of these 18 microRNAs, we selected 14 microRNAs with RQ values lower than or equal to 0.85 (Table 2, column 2, in bold): miR-10b-5p, miR-1185-5p, miR-143-3p, miR-145-5p, miR-199a-3p, miR-23a-3p, miR-26a-5p, miR-29a-3p, miR-30a-5p, miR-335-3p, miR-34a-5p, miR-638, miR-663 and miR-7-5p.
3.3. Validated and Predicted Target Genes
The miRWalk database allowed the identification of validated target genes involved in senescence and extracellular matrix homeostasis, in correlation with the microRNAs of interest (Table 2, column 3). Ingenuity® Pathway Analysis was used to identify new potentially modulated target genes (Table 2, column 5). The genes identified in common with the two methods are also indicated in Table 2, column 4.
Interestingly, among the target genes, family members of collagens (COL1A1, COL2A1, COL2A2, COL3A1, COL4A1, COL 4A2, COL5A3, COL11A2, COL15A1), growth factors and their receptors (EGF, EGFR, VEGFA), transcription factors (SOX2, SOX6, SOX9; FOXO3) or metalloproteinases and their inhibitors (MMP3, MMP9, MMP14; TIMP2) were identified. These genes are well known to be closely related to extracellular matrix homeostasis. Other target genes, such as SIRT1 (Sirtuin 1), SIRT2, BIRC5 (Survivin), AURKB, SHH and LIN28A may also be involved in the fibroblast aging process.
However, miR-1185-5p, miR-23a-3p and miR-335-3p showed no validated target genes in miRWalk, reducing to 11 the list of microRNAs of interest (miR-10b-5p, miR-143-3p, miR-145-5p, miR-199a-3p, miR- 26a-5p, miR-29a-3p, miR-30a-5p, miR-34a-5p, miR-638, miR-663, miR-7-5p) (Table 2, column 1, underlined). The hypothetical effect of the down-regulation of these specific microRNAs on the expression of the potential target gene identified with the miRWalk database is represented in Figure 1. The tendency of a decreased microRNA level observed in our experiments may, as a consequence, induce the up-regulation of the potential target genes listed for each microRNA.
Among the microRNAs, miR-145-5p and miR29a-3p target a large number of genes of interest (respectively, 9 and 10 genes). Some microRNAs (miR-145-5p, miR-26a-5p, miR-29a-3p, miR-30a-5p, miR-34-5p and miR-638) can target collagens (COL1A1, COL2A1, COL2A2, COL3A1, COL4A1, COL 4A2, COL5A3, COL11A2, COL15A1). Interestingly, miR-26a-5p and miR-29a-3p seem to have a high potential for modulating the extracellular matrix composition since they target 3 and 6 collagen family members, respectively (Figure 1). Among the 11 microRNAs studied, miR-26a-5p and miR-335-3p showed a noticeably higher down-regulation following ingredient treatment, with RQ values of 0.577 and 0.573, respectively.
SIRT1, a gene involved in the regulation of cellular senescence, is a miRWalk-validated potential target for miR-34a-5p. Additionally, the Ingenuity® Pathway Analysis predicted that miR-29a-3p and miR-30a-5p may also target SIRT1 and up-regulate its expression, in response to down-regulation (Figure 1).
Genes other than collagens and SIRT1 were targeted by several microRNAs (e.g., SOX2 and VEGFA by 6 microRNAs; SOX9, EGF and EGFR by 3 microRNAs), suggesting the potential importance of these genes in the control of fibroblast homeostasis, extracellular matrix secretion and dermal aging. On the other hand, miR199a-3p and miR-663 are predicted to potentially target only one gene (SOX9 and VEGF1, respectively). The down-regulation of all these microRNAs by the tested ingredient may have a positive impact on extracellular matrix secretion by fibroblasts.
3.4. Interaction Network Representing the Relationship between microRNAs and Potential Targets
Bioinformatics was used to build an overview of the potential interactions between the 18 modulated microRNAs and their potential target genes identified using the miRWalk database. By using Ingenuity® Pathway Analysis, we developed an interaction network (Figure 2(a)). This approach allowed us to further identify 8 new potential target genes closely related to the molecules already included in the network (ADAMTS6, CMA1, COL4A5, FGF13, ITGB6, ITGB8, NID1, RECK), with a potential role in extracellular matrix homeostasis. This interaction network offers the possibility of visualizing all the partners and their interconnections at once. However, 3 microRNAs (miR-10b-5p, miR-335-3p, miR-542-5p) could not be connected to the network. The resulting network without the relationships with microRNAs is depicted in Figure 2(b) to facilitate visualization. This simplified network focuses on the interactions between the 35 miRWalk potential target genes and the 8 new targets identified by using Ingenuity® Pathway Analysis. The addition of new molecules to the network widens the range of targets that may be studied and validated in further in vitro studies.
4. Discussion
Specific “senescence-associated” microRNAs (SA-microRNAs) have emerged as key modulators of cellular senescence by targeting the p53-p21 and p16-pRB pathways. Among these microRNAs, miR-29, miR-30 and
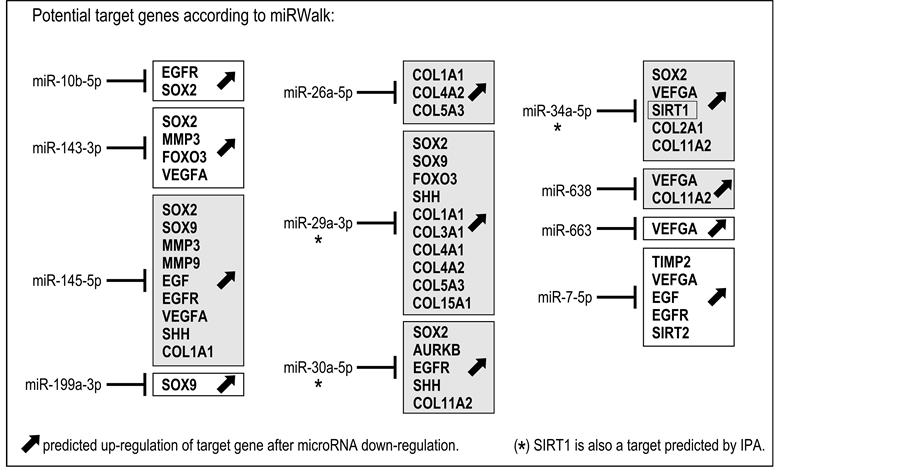
Figure 1. Predictive results on gene expression based on miRWalk-validated target genes. Fibroblasts treated with the tested ingredient expressed 14 microRNAs with RQ ≤ 0.85 (miR-10b-5p, miR-143-3p, miR-145-5p, miR-1185-5p, miR-199a-3p, miR-23a-3p, miR-26a-5p, miR-29a-3p, miR-30a-5p, miR- 34a-5p, miR-335-3p, miR-638, miR-663, miR-7-5p). As miR-1185-5p, miR-23a-3p and miR-335-3p showed no validated target genes in miRWalk, the list of microRNAs of interest was reduced to 11. Predicted up-regulation of potential target genes identified in the miRWalk database is indicated for each of the 11 microRNAs (see also Table 2). Gray boxes indicate predicted up-regulation of collagens as a possible consequence of the down-regulation of miR-145-5p, miR-26a-5p, miR-29a-3p, miR-30a-5p, miR-34a-5p and miR-638. SIRT1 (boxed) is a miRWalk- validated potential target for miR-34a-5p only. However, Ingenuity® Pathway Analysis indicates the possible up-regulation of SIRT1 in response to miR-29a-3p and miR-30a-5p down-regulation (*) (see also Table 2).
miR-34 families are particularly important [23] .
The present study investigated for the first time the modulation of a series of selected microRNAs by a specific complex of ingredients with proven ability to protect human skin cells from senescence in vitro. Results pointed out the regulation of a signature of eleven microRNAs including miR-29a-3p, miR-30a-5p and miR-34a-5p, which are key players in the microRNA-gene regulatory network associated with aging [5] [6] .
The miR-34 family is highly conserved across various organisms and is considered as a tumor suppressor gene that participates in the inhibition of cell cycle progression and the stimulation of apoptosis [24] . Moreover, a recent study has also revealed its role in regulating the lifespan of Caenorhabditis elegans [25] . Interestingly, the accumulation of microRNAs of the miR-34 family with age is also observed in organs or tissues, including the liver [26] , brain [27] and heart [28] . Accordingly, the observed down-regulation of miR-34a-5p by the complex in our study is consistent with the results found in other fields mentioned above. The main mechanism by which the miR-34 family of microRNAs regulates aging or cell senescence, has been elucidated and corresponds to a feedback loop between p53, miR-34 and SIRT1 [29] . Indeed, transcription of the miR-34 family can be directly activated through p53, one of the main factors involved in the inhibition of tumor development and cell
senescence. In turn, miR-34 targets SIRT1, which is consequently inhibited. Given the compelling evidence of the pro-longevity role of SIRT1 in mammals due to its protective effect against cellular oxidative stress, DNA damage and metabolic disturbance [30] , it is reasonable that inhibition of this gene leads to cell senescence and organism aging. In the present study, the observed down-regulation of miR-34a-5p by the complex predicts the possibility of subsequent up-regulation of SIRT1 (Figure 1). Interestingly, aside from miR-34a-5p and according to predictions (Table 2), miR-29a-3p and miR-30a-5p may also regulate SIRT1 (Figure 1; Figure 2(a), red arrows).
The miR-29 family has also been associated with skin aging and senescence. It has been suggested that the miR-29 family plays a pivotal role in the regulation of cell proliferation, extracellular matrix composition, viability
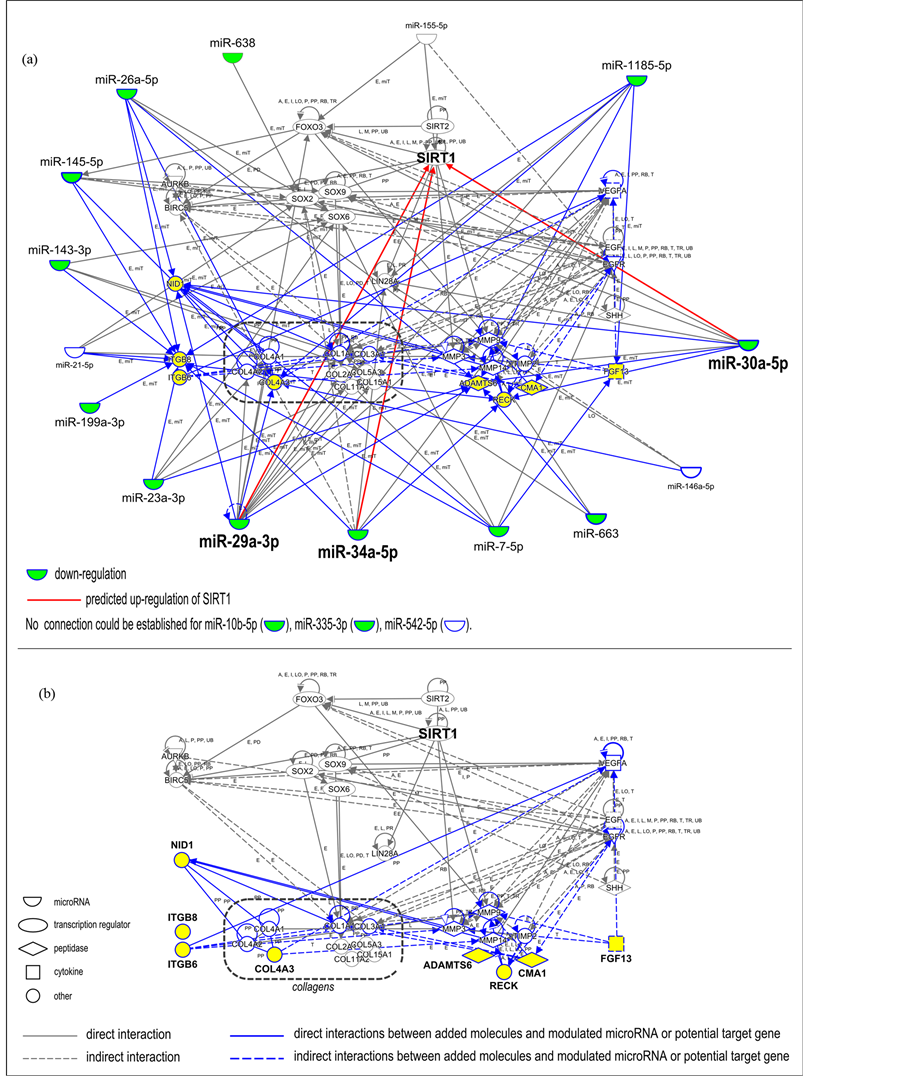
Figure 2. Identification of new genes closely linked with microRNAs modulated by ingredient treatment. (a) Interaction network including i) 18 microRNAs experimentally modulated by the tested ingredient (Table 1), ii) their target genes described in the miRWalk database (Table 2), and iii) 8 new components (highlighted in yellow) identified by building an interaction network with the use of Ingenuity® Pathway Analysis around previously identified microRNAs and genes. Blue lines highlight new relationships that could be established by the introduction of 8 new components in the network. Green color indicates the 14 down-regulated microRNAs with RQ values lower than or equal to 0.85, after application of ingredient complex to fibroblasts. Three microRNAs could not be linked to the network (miR-10b-5p, miR-335-3p, and miR-542-5p). Predicted possible up-regulation of SIRT1 in response to miR-29a-3p, miR-30a-5p and miR-34a-5p down-regulation is indicated (red arrow). (b) Partial interaction network focusing on the relationship between ii) the target genes described in the miRWalk database and iii) the 8 new components of potential interest. Key for the relationship labels and types are available online (http://ingenuity.force.com/ipa/articles/Feature_Description/Legend).
and senescence by controlling multiple overlapping pathways [31] . While some studies point out that the transcriptional activation of the miR-29 family is triggered in response to DNA damage and occurs in a p53-dependent manner in progeroid mice [31] , others demonstrate that miR-29 can negatively regulate B-Myb expression and modulate p16/Rb-driven cellular senescence [19] . Moreover, miR-29a, b, and c were found to be up-regulated during induced and replicative senescence in fibroblasts and are potent repressors of collagen synthesis [32] . It is also important to note that most of the predicted target genes of the miR-29 family encode varied types of collagens, which may account for ECM modulation during skin aging [21] . In our study, the observed down- regulation of miR-29a-3p by the complex is consistent with the predicted positive modulation of collagen expression in the treated fibroblasts (Figure 1). Interestingly, several microRNAs including miR-29a-3p, miR- 30a-5p and miR-34a-5p, appear as possible modulators of collagens (miR-145-5p, miR-26a-5p, miR-29a-3p, miR-30a-5p, miR-34a-5p, and miR-638), reinforcing the possibility of an expected positive modulation on collagen synthesis.
Together with miR-29, miR-30 is also regulated in cellular senescence [19] . Indeed, both miR-29 and miR-30 microRNA families are up-regulated during induced and replicative senescence in vitro. Moreover, interference with miR-29 and miR-30 expression inhibited cellular senescence, strongly suggesting that the level of these microRNAs is correlated with the cellular senescence status [19] . Besides this direct correlation, miR-29 and miR-30 families of microRNAs are also described as being involved in the regulation of ECM synthesis by fibroblasts [21] .
Taken together, our findings suggest that a pattern of microRNAs, including miR-29a-3p, miR-30a-5p and miR-34a-5p, was modulated after the application of 1% of the ingredient complex for 48 h to human skin fibroblasts in culture, and may contribute to the already described activity of the ingredient complex in vitro. Indeed, these microRNAs (miR-10b-5p, miR-143-3p, miR-145-5p, miR-199a-3p, miR-26a-5p, miR-29a-3p, miR-30a-5p, miR-34a-5p, miR-638, miR-663, miR-7-5p) may play a role in the observed protection against cellular senescence in vitro, and UV-induced DNA damage [13] - [16] , by the possible regulation of collagen and SIRT1 expression levels. However, further validation is required in order to link microRNA down-regulation and effective up-regulation of the predicted target genes.
Acknowledgments
We express our appreciation for the valuable discussions with Noëlle Garcia, Audrey Le Mestr, Laura Restellini, Magali Bonnans and Rachel Chabert.
Cite this paper
XianghongYan,CatherineSerre,LaurineBergeron,LudivineMur,ValèreBusuttil,Jean-MarieBotto,NouhaDomloge, (2015) Modulation of a Specific Pattern of microRNAs, Including miR-29a, miR-30a and miR-34a, in Cultured Human Skin Fibroblasts, in Response to the Application of a Biofunctional Ingredient that Protects against Cellular Senescence in Vitro. Journal of Cosmetics, Dermatological Sciences and Applications,05,332-342. doi: 10.4236/jcdsa.2015.54040
References
- 1. Jung, H.J. and Suh, Y. (2012) MicroRNA in Aging: From Discovery to Biology. Current Genomics, 13, 548-557.
http://dx.doi.org/10.2174/138920212803251436 - 2. Smith-Vikos, T. and Slack, F.J. (2012) MicroRNAs and Their Roles in Aging. Journal of Cell Science, 125, 7-17.
http://dx.doi.org/10.1242/jcs.099200 - 3. Botto, J.M., Busuttil, V., Labarrade, F., Serre, C., Bergeron, L., Capallere, C. and Domloge, N. (2015) MicroRNAs in Skin Physiology. In: Gadberry, R.J., Epstein, H. and Botto, J.-M., Eds., Cosmetic Industry Approaches to Epigenetics and Molecular Biology (Harrys Cosmeticology 9th Edition), Chemical Publishing Company, Revere, MA, 940-1189.
- 4. Engels, B.M. and Hutvagner, G. (2006) Principles and Effects of microRNA-Mediated Post-Transcriptional Gene Regulation. Oncogene, 25, 6163-6169.
http://dx.doi.org/10.1038/sj.onc.1209909 - 5. Zhang, X., Azhar, G. and Wei, J.Y. (2012) The Expression of microRNA and microRNA Clusters in the Aging Heart. PLoS One, 7, e34688.
http://dx.doi.org/10.1371/journal.pone.0034688 - 6. Zhao, X., Huang, Y., Wang, Y., Chen, P., Yu, Y. and Song, Z. (2013) MicroRNA Profile Comparison of the Corneal Endothelia of Young and Old Mice: Implications for Senescence of the Corneal Endothelium. Molecular Vision, 19, 1815-1825.
- 7. Kwekel, J.C., Vijay, V., Desai, V.G., Moland, C.L. and Fuscoe, J.C. (2015) Age and Sex Differences in Kidney microRNA Expression during the Life Span of F344 Rats. Biology of Sex Differences, 6, 1.
http://dx.doi.org/10.1186/s13293-014-0019-1 - 8. Zhang, L., Li, J. and Zhou, K. (2009) Chelating and Radical Scavenging Activities of Soy Protein Hydrolysates Prepared from Microbial Proteases and Their Effect on Meat Lipid Peroxidation. Bioresource Technology, 101, 2084-2089.
http://dx.doi.org/10.1016/j.biortech.2009.11.078 - 9. Imbert, I., Botto, J.M., Dal Farra, C. and Domloge, N. (2012) Modulation of Telomere Binding Proteins: A Future Area of Research for Skin Protection and Anti-Aging Target. Journal of Cosmetic Dermatology, 11, 162-166.
http://dx.doi.org/10.1111/j.1473-2165.2012.00611.x - 10. De Lange, T. (2005) Shelterin: The Protein Complex That Shapes and Safeguards Human Telomeres. Genes & Development, 19, 2100-2110.
http://dx.doi.org/10.1101/gad.1346005 - 11. Stewart, J.A., Chaiken, M.F., Wang, F. and Price, C.M. (2012) Maintaining the End: Roles of Telomere Proteins in End-Protection, Telomere Replication and Length Regulation. Mutation Research, 730, 12-19.
http://dx.doi.org/10.1016/j.mrfmmm.2011.08.011 - 12. Bergeron, L., Lebleu, A., Court, E., Busuttil, V., Botto, J.M. and Domloge, N. (2012) Modulating TRF2 Expression Preserves Telomere Integrity and Prevents DNA DSBs, during Aging. Journal of Investigative Dermatology, 132, S51-S64.
- 13. Dweep, H., Sticht, C., Pandey, P. and Gretz, N. (2011) miRWalk—Database: Prediction of Possible miRNA Binding Sites by “Walking” the Genes of Three Genomes. Journal of Biomedical Informatics, 44, 839-847.
http://dx.doi.org/10.1016/j.jbi.2011.05.002 - 14. Vlachos, I.S., Paraskevopoulou, M.D., Karagkouni, D., Georgakilas, G., Vergoulis, T., Kanellos, I., Anastasopoulos, I.L., Maniou, S., Karathanou, K., Kalfakakou, D., Fevgas, A., Dalamagas, T. and Hatzigeorgiou, A.G. (2015) DIANA-TarBase v7.0: Indexing More than Half a Million Experimentally Supported miRNA:mRNA Interactions. Nucleic Acids Research, 43, D153-D159.
http://dx.doi.org/10.1093/nar/gku1215 - 15. Xiao, F., Zuo, Z., Cai, G., Kang, S., Gao, X. and Li, T. (2009) miRecords: An Integrated Resource for MicroRNA-Target Interactions. Nucleic Acids Research, 37, D105-D110.
http://dx.doi.org/10.1093/nar/gkn851 - 16. Lewis, B.P., Burge, C.B. and Bartel, D.P. (2005) Conserved Seed Pairing, Often Flanked by Adenosines, Indicates That Thousands of Human Genes Are MicroRNA Targets. Cell, 120, 15-20.
http://dx.doi.org/10.1016/j.cell.2004.12.035 - 17. Bonifacio, L. and Jarstfer, M. (2010) miRNA Profile Associated with Replicative Senescence, Extended Cell Culture, and Ectopic Telomerase Expression in Human Foreskin Fibroblasts. PLoS ONE, 5, e12519.
http://dx.doi.org/10.1371/journal.pone.0012519 - 18. Marasa, B., Srikantan, S., Martindale, J., Kim, M.M., Lee, E.K., Gorospe, M. and Abdelmohsen, K. (2010) MicroRNA Profiling in Human Diploid Fibroblasts Uncovers miR-519 Role in Replicative Senescence. Aging, 2, 333-343.
- 19. Martinez, I., Cazalla, D., Almstead, L., Steitz, J.A. and DiMaio, D. (2011) miR-29 and miR-30 Regulate B-Myb Expression during Cellular Senescence. Proceedings of the National Academy of Sciences of the United States of America, 108, 522-527.
http://dx.doi.org/10.1073/pnas.1017346108 - 20. Maes, O.C., Sarojini, H. and Wang, E. (2009) Stepwise Up-Regulation of MicroRNA Expression Levels from Replicating to Reversible and Irreversible Growth Arrest States in WI-38 Human Fibroblasts. Journal of Cellular Physiology, 221, 109-119.
http://dx.doi.org/10.1002/jcp.21834 - 21. Suh, E.J., Remillard, M.Y., Legesse-Miller, A., Johnson, E.L., Lemons, J.M., Chapman, T.R., Forman, J.J., Kojima, M., Silberman, E.S. and Coller, H.A. (2012) A MicroRNA Network Regulates Proliferative Timing and Extracellular Matrix Synthesis during Cellular Quiescence in Fibroblasts. Genome Biology, 13, R121.
http://dx.doi.org/10.1186/gb-2012-13-12-r121 - 22. Tomé, M., Lopez-Romero, P., Albo, C., Sepúlveda, J.C., Fernández-Gutiérrez, B., Dopazo, A., Bernad, A. and González, MA. (2011) miR-335 Orchestrates Cell Proliferation, Migration and Differentiation in Human Mesenchymal Stem Cells. Cell Death & Differentiation, 18, 985-995.
http://dx.doi.org/10.1038/cdd.2010.167 - 23. Olivieri, F., Rippo, M.R., Monsurrò, V., Salvioli, S., Capri, M., Procopio, A.D. and Franceschi, C. (2013) MicroRNAs Linking Inflamm-Aging, Cellular Senescence and Cancer. Ageing Research Reviews, 12, 1056-1068.
http://dx.doi.org/10.1016/j.arr.2013.05.001 - 24. He, L., He, X., Lim, L.P., de Stanchina, E., Xuan, Z., Liang, Y., Xue, W., Zender, L., Magnus, J., Ridzon, D., Jackson, A.L., Linsley, P.S., Chen, C., Lowe, S.W., Cleary, M.A. and Hannon, G.J. (2007) A MicroRNA Component of the p53 Tumour Suppressor Network. Nature, 447, 1130-1134.
http://dx.doi.org/10.1038/nature05939 - 25. Yang, J., Chen, D., He, Y., Meléndez, A., Feng, Z., Hong, Q., Bai, X., Li, Q., Cai, G., Wang, J. and Chen, X. (2013) miR-34 Modulates Caenorhabditis elegans Lifespan via Repressing the Autophagy Gene atg9. AGE, 35, 11-22.
http://dx.doi.org/10.1007/s11357-011-9324-3 - 26. Li, N., Muthusamy, S., Liang, R., Sarojini, H. and Wang, E. (2011) Increased Expression of miR-34a and miR-93 in Rat Liver during Aging, and Their Impact on the Expression of Mgst1 and Sirt1. Mechanisms of Ageing and Development, 132, 75-85.
http://dx.doi.org/10.1016/j.mad.2010.12.004 - 27. Liu, N., Landreh, M., Cao, K., Abe, M., Hendriks, G.J., Kennerdell, J.R., Zhu, Y., Wang, L.S. and Bonini, N.M. (2012) The MicroRNA miR-34 Modulates Ageing and Neurodegeneration in Drosophila. Nature, 482, 519-523.
http://dx.doi.org/10.1038/nature10810 - 28. Seeger, T. and Boon, R.A. (2015) MicroRNAs in Cardiovascular Aging. The Journal of Physiology.
http://dx.doi.org/10.1113/JP270557 - 29. Yamakuchi, M. and Lowenstein, C.J. (2009) miR-34, SIRT1 and p53: The Feedback Loop. Cell Cycle, 8, 712-715.
http://dx.doi.org/10.4161/cc.8.5.7753 - 30. Lopez-Otin, C., Blasco, M.A., Partridge, L., Serrano, M. and Kroemer, G. (2013) The Hallmarks of Aging. Cell, 153, 1194-1217.
http://dx.doi.org/10.1016/j.cell.2013.05.039 - 31. Ugalde, A.P., Espanol, Y. and Lopez-Otin, C. (2011) Micromanaging Aging with miRNAs: New Messages from the Nuclear Envelope. Nucleus, 2, 549-555.
http://dx.doi.org/10.4161/nucl.2.6.17986 - 32. Feliciano, A., Sánchez-Sendra, B., Kondoh, H. and Lleonart, M.E. (2011) MicroRNAs Regulate Key Effector Pathways of Senescence. Journal of Aging Research, 2011, Article ID: 205378.
NOTES

*Corresponding author.


