Advances in Bioscience and Biotechnology
Vol.3 No.4(2012), Article ID:21862,9 pages DOI:10.4236/abb.2012.34051
Characteristics of dehalogenase from bacteria isolated from the gut of pond-reared rohu (Labeo rohita) juveniles in Myanmar
![]()
1Faculty of Biosciences and Bioengineering, Universiti Teknologi Malaysia, Johor Bahru, Malaysia
2Fish Health Section, Aquaculture Department, Southeast Asian Fisheries Development Center, Iloilo, Philippines
3Fish Health Laboratory, Department of Fisheries, Yangon, Myanmar
Email: *fzhutm@gmail.com
Received 24 May 2012; revised 28 June 2012; accepted 4 July 2012
Keywords: 2,2-dichloropropionic acid; 16S rDNA; Labeo rohita; dehalogenase gene; biodegradation; haloalkanoic acid; dichloropropionate
ABSTRACT
Unwarranted accumulation of halogenated compounds in the rivers and streams has in recent years emerged due to the widespread use agricultural pesticides. The presence of these halogenated compounds in the water does not only suppress the immune system of fish but adversely induce serious morbidity and mortality among cultured stocks. Importantly, gradual accumulation of these compounds in the system of cultured and wild freshwater fish species cultured in ponds and floating net-cages in dams and rivers, respectively, poses some risks to humans, the end users. In this study, we attempted to isolate bacteria from the gut of pond-reared rohu (Labeo rohita) in Myanmar, screened the isolated bacteria for dehalogenase gene using molecular technique and tested the ability of these bacteria to degrade halogenated compounds in vitro. The eight bacterial strains studied were identified as Enterobacter mori strain MK-121001, Enterobacter cloacae strains MK121003, MK-121004, MK121010, Ralstonia solanacearum strain 121002, Acinetobacter baumannii strain MK121007, Chromobacterium violaceum strain MK121009 and Pantoea vagans strain 121011. Only three bacterial strains (MK121002, MK121007 and MK121009) were capable of degrading 2,2-dichloropropionic acid (2,2-DCP) as the sole carbon source up to a final substrate concentration of 20 mM. Their mean growth doubling time ranging from 6 - 23 hours with the maximum of chloride ion released of 85%. PCR amplification with oligonucleotide primers designed from group I dehalogenase revealed the presence of dehalogenase genes in all isolates suggesting dehalogenase gene in strains 121001, 121003, 121004, 121010 and 121011 were silenced. In contrast, group II dehalogenase primers did not show any PCR amplification. These results suggest that MK121002, MK121007 and MK121009 only encode a group I dehalogenase and its non-stereoselectivity is in agreement with previoulsly described group I haloacid dehalogenase. The partial gene sequences were blasted but no significant sequence identity was observed. Therefore, it suggests the 2-haloacid dehalogenase of MK121002, MK12-1007 and MK- 121009 might be a novel group I 2-haloacid dehalogenase. The results indicated a broad distribution of dehalogenation genes in many microbial genomes that harbor dehalogenase(s) due to the exposure of the microorganisms to the naturally occurring or man-made halogenated compounds in the environmental systems. So far, microorganisms capable of producing dehalogenases were mainly isolated from soil and scarcely from aquatic animals and their environments. To the authors’ knowledge, this is the first report on the isolation of dehalogenase-producing bacteria from the gut of pond-reared freshwater fish, Labeo rohita, in Myanmar.
1. INTRODUCTION
Halogenated compounds are widely used as herbicides, pesticides, insecticides and antibiotics. These chemicals are produced in large quantities by the chemical industry. Extensive use of herbicides for example, led to harmful effects on human and natural environment because many synthetic organohalogens are generally persistent, bioaccumulative and toxic. Halo-aliphatic compounds such as 2,2-dichloropropionic acid (2,2-DCP) also known as Dalapon, is commonly used as herbicides. Its presence in the environment is toxic and degradation of this compound by microorganisms is of great interest in bioremediation [1]. Degradation of herbicide Dalapon was reported earlier by Magee and Colmer [2] after observation of bacteria that produce dehalogenase enzyme. Since then, studies on isolation of microbes that potentially produce dehalogenase have been undertaken [3-6]. Several bacteria are known to be able to degrade these haloaliphatic pollutants and many of these microorganisms have been isolated from the soil contaminated area or some from marine environment [7-10].
The study of dehalogenases become apparent based on two aspects, first to understand the diversity of α or β haloalkanoic dehalogenase [11,12] in terms of the origin of novel enzyme activities and the adaption of bacteria to degrade xenobiotic compunds. Second, some of the isolated microorganism posses multiple dehalogenases for example in Rhizobium sp. RC1 [13] and Pseudomonas putida PP3 [14] suggesting the complexity of the control of dehalogenase gene regulation [15]. Therefore, studies on newly identified dehalogenases may relate to these two important areas. In addition, hydrolytic dehalogenases are popular for research because they are cytosolic proteins and do not require cofactors and use water as the co-substrate [16-18].
The rohu, Labeo rohita, is an herbivorous fish that inhabits the tropical lowland river systems of Pakistan, northern India and Myanmar. Rohu is considered to be the most important of the Indian major carps and is the world’s 10th highest cultured finfish by production volume in India [19]. Rohu is a eurythermal species and does not thrive at temperatures below 14˚C. It is a fast growing species and attains about 35 - 45 cm total length and 700 - 800 g in one year under normal culture conditions. As the gut of fish is an open system, it harbors microbial populations from the aquatic environment through water and food which are populated with bacteria. Water pollution with chemical contaminants has become one of the most crucial environmental issues of the century. Therefore, the freshwater ecosystems are confronted with high levels of xenobiotic chemicals due to the pollutants transported from industrial areas into the environment [20]. Among the freshwater fishes, carps are most affected to environmental contamination [21,22].
Dehalogenase producing microorganisms have been frequently isolated from soil and marine environment but none from other animals. So far, there is no study that have been reported on the association of pollutant degrading bacteria in the gut of Labeo rohita fish. In the present study focuses on the identification and characterization of potential bacteria from gut of Labeo rohita that able to utilize 2,2-DCP as only carbon source.
2. MATERIALS AND METHODS
2.1. Fish Sample
The fish Labeo rohita were collected from the fish farm located in the remote area in Myanmar. The fish farm was highly exposed by the surrounding area with the herbicides, pesticides and molluskocides. These substances may have seeping into the pond of the growing fish. The fish were collected from the pond and brought to the lab. Using aseptic technique, the fish gut was cut open for bacterial isolation.
2.2. Chemicals
Halogenated compounds—Monochloroacetic acid (MCA), Dichloroacetic acid (DCA), Trichloroacetic acid (TCA), D,L-2-chloropropionic acid (D,L-2-CP), D-2-chloropropionic acid (D-2-CP), L-2-chloropropionic acid (L-2-CP), 3-chloropropionic acid (3-CP), 2,2-dichloropropionic acid (2,2-DCP), 2-chlorobutyric acid, 3-chlorobutyric acid and 2,2,3-trichlorobutyric acid were obtained from SigmaAldrich Chemical Co. (USA). Other chemicals were of analytical grade.
2.3. Growth Conditions
The bacterial culture was inoculated into 100 ml of minimal media containing 2,2-DCP as the sole source of carbon and energy. The flasks were incubated at 30˚C in a rotary incubator at 150 rpm. The liquid minimal media employed for growth consisted of 10× concentration basal salts containing K2HPO4∙3H2O (42.5 g∙L−1), NaH2PO4∙2H2O (10.0 g∙L−1) and (NH4)2∙SO4 (20.0 g∙L−1). The trace metals salts solution was prepared at 10x concentrate that contained nitriloacetic acid (NTA) (1.0 g∙L−1), MgSO4∙7H2O (2.0 g∙L−1), FeSO4∙7H2O (120.0 mg∙L−1), MnSO4∙4H2O (30.0 mg∙L−1), ZnSO4∙7H2O (30.0 mg∙L−1) and CoCl2∙6H2O (10.0 mg∙L−1) in distilled water [23]. The minimal media for growing bacteria contained 10 ml of 10× basal salts and 10 ml of 10× trace metal salts per 100 ml of distilled water were autoclaved (121˚C for 15 min at 15psi). The carbon sources MCA, DCA, TCA, D,L-2-CP, D-2-CP, L-2-CP, 3-CP, 2,2-DCP, 2-chlorobutyric acid, 3-chlorobutyric acid and 2,2,3-trichlorobutyric acid were sterilized by filtration separately and added aseptically to the media to desired final concentration of 10 and/or 20 mM. In order to prepare solid medium, Oxoid bacteriological agar (1.5% w/v) was added prior to sterilization. For the isolation of pure colonies, 0.1 mL of aliquot was spread onto solid minimal media contained 20 mM 2,2-DCP and incubated at 30˚C.
2.4. DNA Extraction and Amplification of the 16S rDNA Gene
Bacterial DNA was extracted from bacterial cultures grown on 20 mM 2,2-DCP minimal media using Wizard® Genomic DNA Purification Kit (Promega). The polymerase chain reaction (PCR) was carried out to amplify the 16S rDNA gene and the universal primers used Fd1 (5’-AGA GTT TGA TCC TGG CTC AG-3’) and rP1 (5’-ACG GTC ATA CCT TGT TAC GAC TT-3’) [24]. The amplification reactions contained in 50 µl with template DNA, forward primer (Fd1), reverse primer (rP1), PCR master mix (Fermentas Inc. USA) and nuclease free water. DNA amplification was performed for 30 cycles and the PCR cycle was set as initial denaturation 94˚C for 5 min, followed by cooling, denaturation 94˚C for 1 min, annealing 55˚C for 1 min and final extension 72˚C for 10 min.
2.5. Amplification of Putative Dehalogenase Gene Using Degenerate Primers
Primers dehIfor1 (5’-ACGY TNSG SGTG CCNT GGGT-3’) and dehIrev2 (5’-SGCM AKSR CNYK GWAR TCACT-3’) were adapted from group I dehalogenase; Primers dehIIfor1 (5’-TGG CGV CAR MRD CAR CTB GAR TA-3’) and dehIIrev1 (5’-TCSM ADSB RTTB GAS GAN ACR AA-3’) were adapted from group II dehalogenase [11]. These primers were designed on the basis of consensus dehalogenase derived from an alignment of dehalogenase genes belongs to the same group although the identification of genes using a sequence-based method is challenging due to low sequence conservation among dehalogenases. PCR amplification was performed as described by Hill et al. [11]. PCR products were visualized by agarose electrophoresis [25].
2.6. DNA Sequencing of Partial Dehalogenase Gene and Analysis
PCR products were sent for sequencing at First BASE Laboratories (Malaysia) Sdn. Bhd. DNA was purified using Qiagen PCR purification Kit. Both DNA strands were sequenced to ensure accuracy. The dehalogenase gene sequences were compared with those in the GenBank-EMBL/NCBI database.
2.7. Partial Biochemical Test
Several of biochemical tests were carried out including indole test, oxidase test, reduction of nitrates to nitrites, acid fast stain, gelatin hydrolysis, sucrose test, citrate test and catalase test to characterize bacteria’s biochemical properties.
2.8. Determination of the Bacteria Growth and Chloride Ion Released Estimation
The growth pattern of the bacteria culture in 20 mM 2,2-DCP minimal medium were monitored at appropriate time interval by taking 1 ml sample from the growth medium and measured at A600nm. The chloride ion in the growth medium was monitored using 1 ml of the same sample for growth monitoring, then aliquot into 100 μl of 0.25 M ammonium ferric sulphate in 9 M nitric acid and then followed by 100 μl of mercuric thiocyanate. After 10 min incubation at room temperature the reading was measured at A460nm.
2.9. Dehalogenase Enzyme Assay
Cell-free extracts were prepared according to Mesri et al. [12]. Dehalogenase activity was measured by determining the release of chloride ion as described by Bergman and Sanik [26]. Dehalogenase activity was determined as total chloride released at 30˚C in a reaction containing 0.1 M Tris-acetate buffer (pH 7.5) (4700 μl), 1 mM halogenated aliphatic acid (50 μl of a 0.1 M stock), water and enzyme from cell-free extract added to a final volume of 5 ml. All reaction components except the enzyme were combined and allowed to equilibrate for 5 min at 30˚C, then the reaction was initiated by adding cell-free extract. Samples (1.0 ml) were removed at appropriate intervals and assayed for halide ions. Protein was determined by the biuret procedure with crystalline egg albumin as a standard [27]. Specific activity was defined as the micromoles of chloride liberated per milligram protein in 10 min under the stated conditions.
3. RESULTS
3.1. Identification of 2,2-DCP Degrading Bacteria
The bacterial strains isolated from fish were screened on solid minimal media containing 20 mM 2,2-DCP as a carbon source. All 8 potential colonies were selected and further investigated by growing them in 20 mM liquid minimal media with 3 strains were detected to grow well. Strains MK121002 and MK121009 showed 3 to 4 times faster growth than MK121007, with more than 85% maximum chloride ion released detected in the growth minimal medium. Overall growth properties were described in Table 1. However, the cells could not grow in concentration above 20 mM 2,2-DCP due to the toxicity of the substrate used. The morphological and partial biochemical characteristics were summarized in Table 2. Tentative identification of the isolates were deduced according to Bergey’s Manual of Systematic Bacteriology [28]. The bacterial isolates were also identified on the basis of 16S rDNA sequences. Therefore, the isolated bacteria were designated as shown in Table 3.
3.2. Bacterial Growth in Other Halogenated Compounds
All 8 bacterial strains were tested to grow in 10 mM

Table 1. Growth properties of bacterial isolates in 20 mM 2,2-DCP liquid minimal medium at 30˚C on orbital incubator shaker.

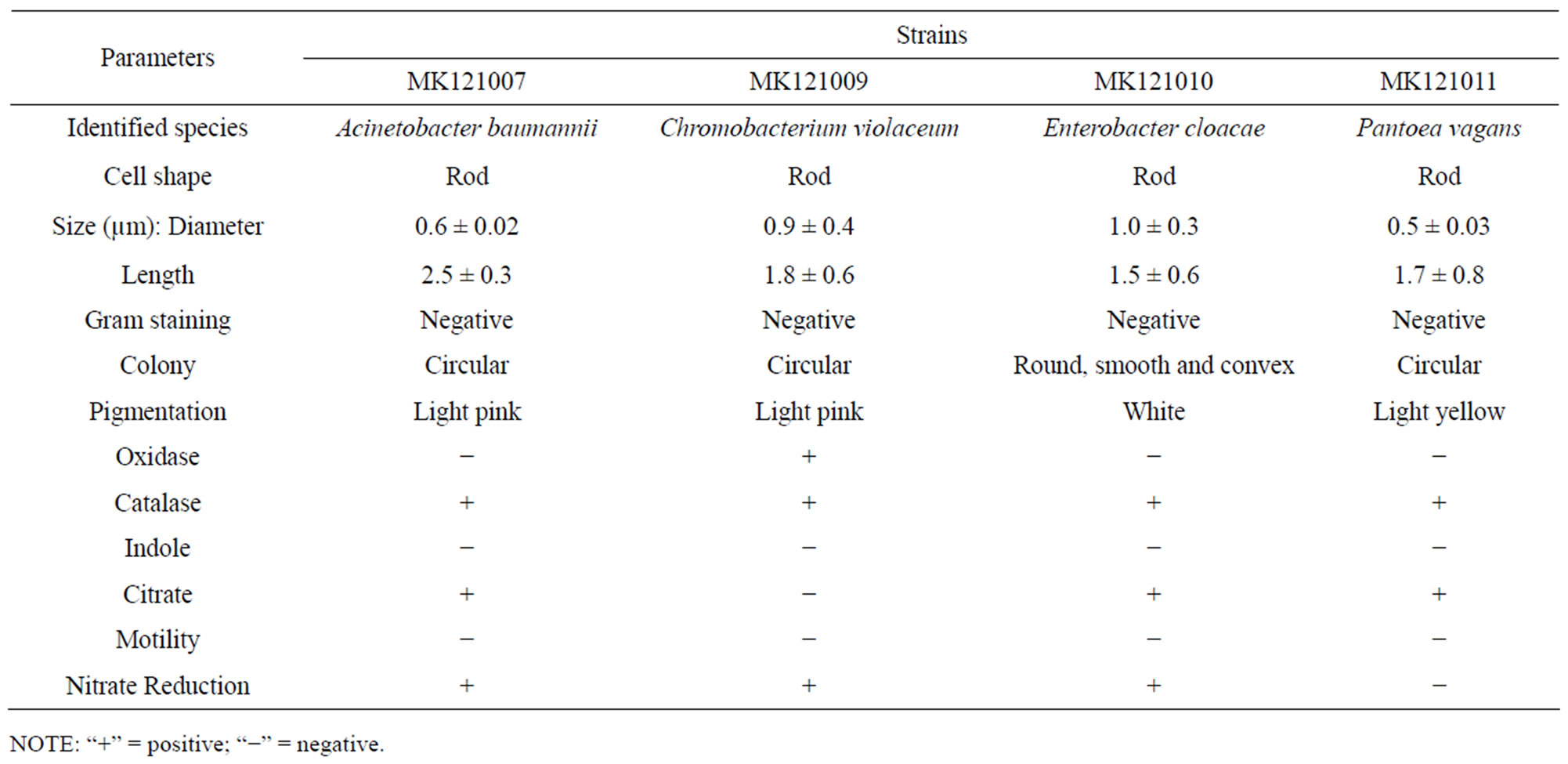
Table 2. Morphological and partial biochemical characteristics of the bacterial isolates.

Table 3. Similarity percentage of 16S rDNA sequence of bacterial strains compared to those obtained from BLAST search.
3-CP and 10 mM 2,3-DCP liquid minimal medium. The results showed all isolates failed to grow in any of these substrates suggesting 3-CP and 2,3-DCP were not substrates for growth.
3.3. PCR Amplification of Dehalogenase Genes and Analysis
Oligonucleotide primers derived from group I dehalogenase were used during PCR reaction using genomic DNA extracted from all of the isolates. PCR fragments of approximately 1 kb fragments were generated. Surprisingly, DNA template from strains 121001, 121003, 121004, 121010 and 121011 were also showed PCR amplification similar to the band pattern to that of strains that can grow well in 2,2-DCP, suggesting non-growth organisms carry the dehalogenase gene that were not expressed or silenced (Figure 1). In contrast, group II dehalogenase primers did not show any PCR amplification. These results suggest that MK121002, MK121007 and MK121009 only encode a group I dehalogenase and its non-stereoselectivity is in agreement with previoulsly described group I haloacid dehalogenase. The partial gene and amino acid sequences were blasted in NCBI database, but no significant sequence identity was observed. These results suggest the 2-haloacid dehalogenase of MK121002, MK121007 and MK121009 might be a novel group I 2-haloacid dehalogenase. The nucleotide sequence of the partial gene has been submitted to NCBI GenBank under accession number JX121567, JX121568 and JX121569.
3.4. Dehalogenase Activity in Cell Free Extracts
Dehalogenase activity was assessed as the liberation of free halide in cell free extracts from bacteria (strain MK121002, MK121007 and MK121009) grown on 20 mM 2,2-DCP as the sole carbon source. Halide liberation was detected from extracts of cells grown in 2,2-DCP only. However, no halide was detected by extracts of the same bacteria grown in the presence of MCA, 3CP or 2-chlorobutyric acid. Extracts of bacteria grown in minimal media containing non-halogenated compound such as lactate, pyruvate or glucose as sole soure of carbon did not exhibit dehalogenase activity as expected. The specificity of the dehalogenase for chlorinated aliphatic acids is shown in Table 4.
4. DISCUSSION
Bacteria taxonomy using 16S rDNA is a common method in the characterization and identification of microorganism. The determination of 16S rDNA gene sequences is a routine procedure in prokaryotic taxonomy, resulting in large and growing databases, which improve phylogeny reconstructions, identification results and primer specificity evaluations [29]. In this paper, we described the isolation and identification of bacterial strains from the gut of Labeo rohita. The 16S rDNA gene sequence analysis confirmed the identity of the organisms (Table 3).
Growth experiment showed good growth at 20 mM 2,2-DCP minimal media. The cells doubling time was calculated as summarized in Table 1. Both strains MK- 121002 and MK121009 were somewhat grow faster than previously reported Rhizobium sp. RC1 growing in the same substrate [13,30]. In contrast, strain MK121007 showed slow growth possibly due to the inefficient uptake of substrate into the cell by permease system [31,32] and can be depicted with low enzyme activity as in Table 4. Apart from growth on 2,2-DCP, all strains were unable to grow on 10 mM 3-CP and 2,3-DCP which differed only in chlorine substitution position. Degradation of 3- CP or β-haloalkanoic acid by microorganisms were rare [12].
Current investigation strongly suggest that gut microorganism like Ralstonia solanacearum strain 121002, Acinetobacter baumannii strain MK121007 and Chromobacterium violaceum strain MK121009 produced dehalogenase enzyme that capable to act on other halogenated compounds but restricted to only propionic group with halide ion located at carbon number two. Other microbes, like Bacillus sp. was isolated from the gut of the Indian major carp (rohu) or Labeo rohita. These strains were identified as B. circulans, B. pumilus and B. cereus [33]. The presence of this bacteria especially in the gastrointestinal tracts of Labeo rohita will help in digestion. Genus Enterobacter and Pantoea were also isolated in current study. Their presence in fish gut was possibly correlated to its feeding habit [34-38]. So far, there is no reported case of the respective bacteria which could degrade halogenated compound.
In previous studies have shown that genus Acinetobacter and Chromobacterium capable of degrading phenol as a
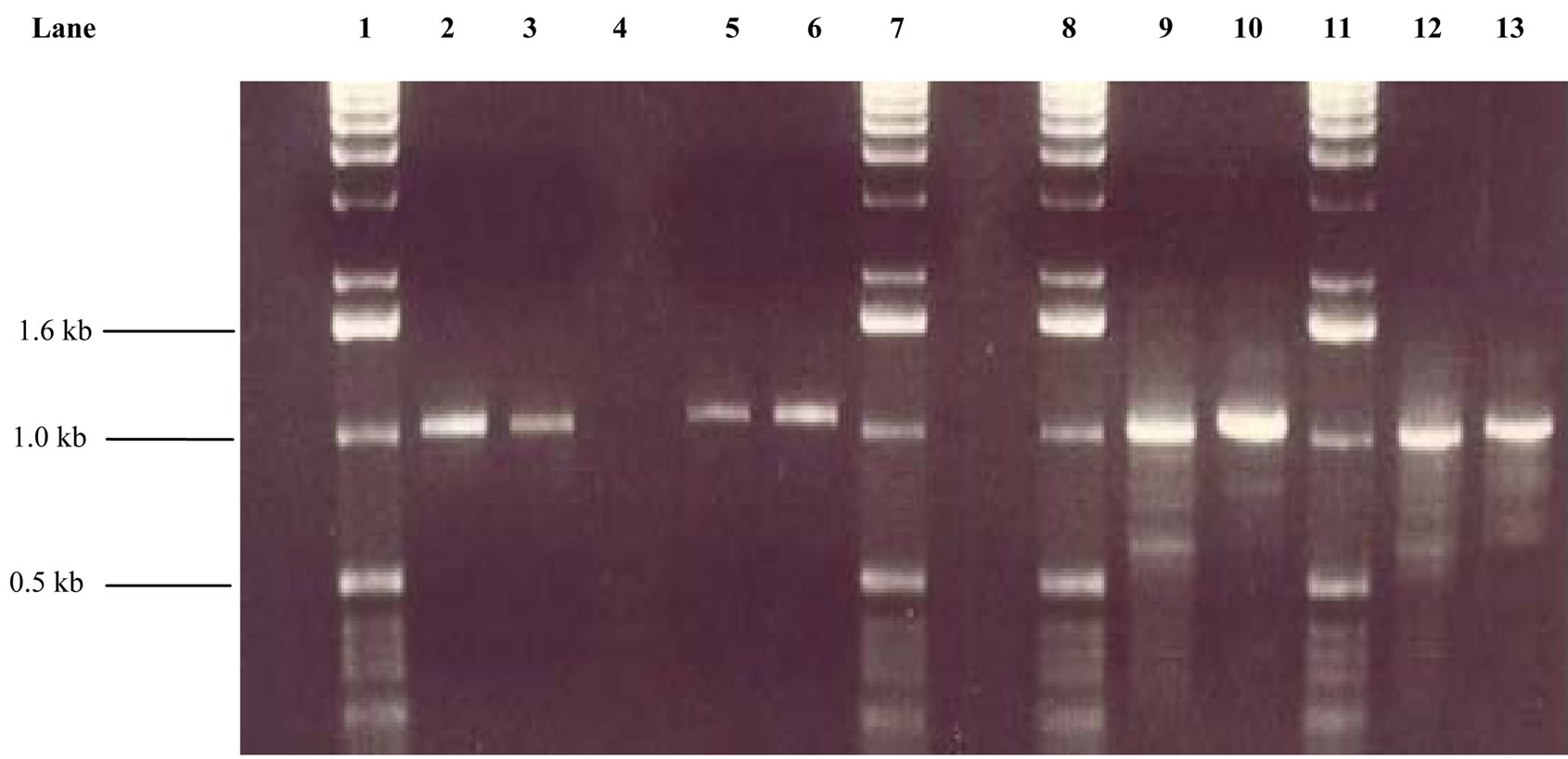
Figure 1. PCR analysis of genomic DNA extracted from the isolated bacteria. Lane 1 = 1kb ladder; Lane 2 = Ralstonia solanacearum MK121002; Lane 3 = Acinetobacter baumannii MK121007; Lane 4 = Control (E. coli) Lane 5 = Enterobacter mori MK121001; Lane 6 = Chromobacterium violaceum MK121009; Lane 7 = 1kb ladder; Lane 8 = 1kb ladder; Lane 9 = Enterobacter cloacae MK121003; Lane 10 = Enterobacter cloacae MK- 121004; Lane 11 = 1 kb ladder; Lane 12 = Enterobacter cloacae MK121010; Lane 13 = Pantoea vagans MK- 121011.
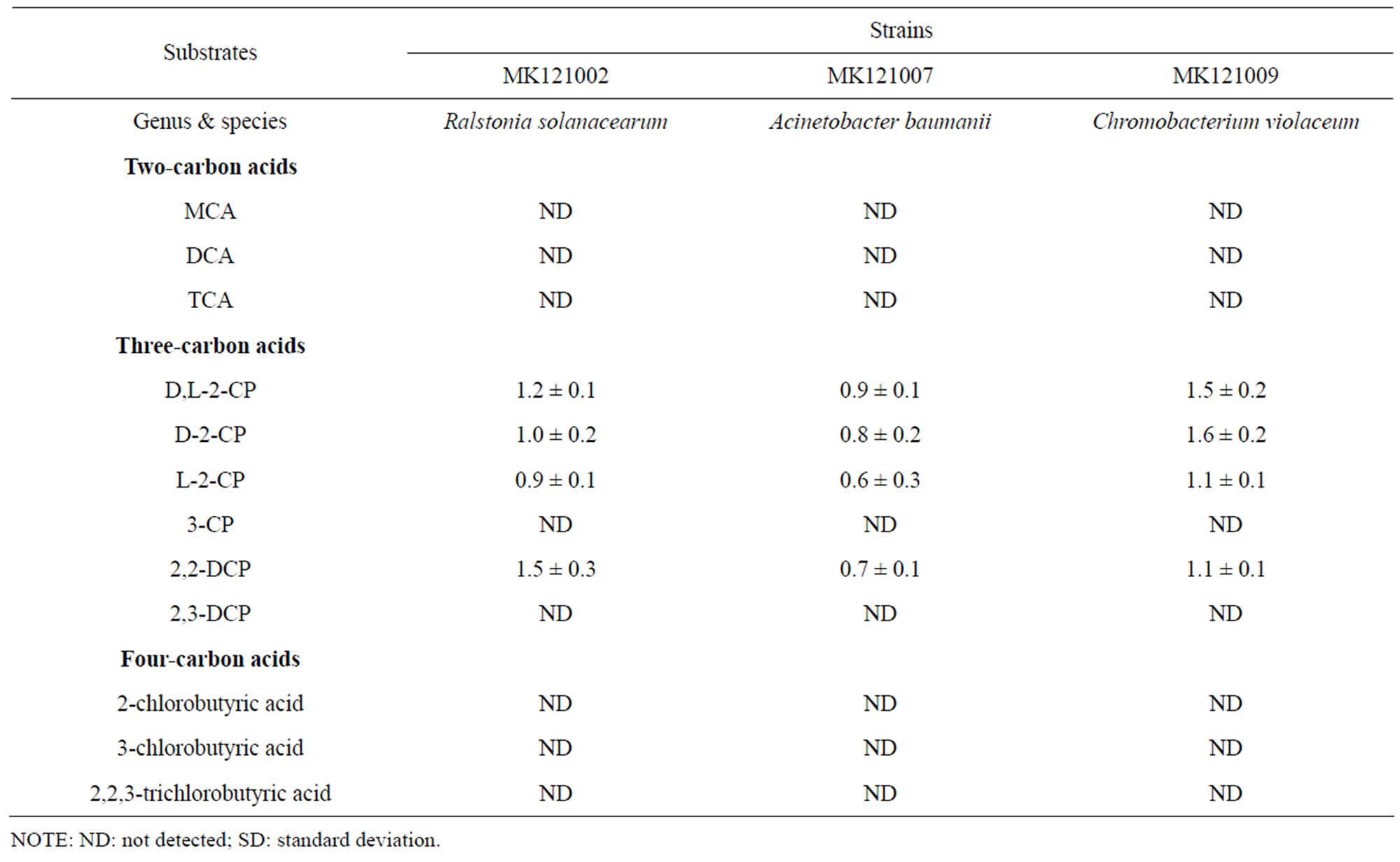
Table 4. Activity of dehalogenase (µmol∙Cl−1∙min−1∙mg−1 protein ± SD) using various substrates.
sole energy and carbon source [39-42]. Other xenobiotic compounds such as toluene [43], 4-hydroxy-benzoate [44] and 2-chloro-N-isopropylacetanilide [45] can be metabolized to their corresponding benzoates by various Acinetobacter strains. According to Mergeay et al. (2003) [46], Ralstonia sp. has been reported to degrade numerous types of hydrocarbons such as polyaromatic hydrocarbons (PAHs) and plays a significant role in degrading various environmental pollutants. In addition to that, Ralstonia eutropha CA-11 isolated from a rice paddy soil was also found to degrade 2,3-DCP, MCA and D, L-2-CP under aerobic conditions [47].
As shown in Figure 1, all isolates had approximately 1.0 kb PCR product with the group I PCR primers, the size of which 50% larger than the previous report [11]. These results suggest that all isolates encodes a group I dehalogenase and its non-stereospecificity is in agreement with previously described group I 2-haloacid dehalogenases [3,48-51]. In addition, the genes amplified from all non-growth organisms on 2,2-DCP suggest the dehalogenase gene in these organisms is silenced. The properties of chd1 gene also known as the silence or cryptic dehalogenase gene was described earlier by Tsang and Sam [52].
In conclusion, this study provides the identity of the bacteria that can grow on halogenated compound. However, some of the identified bacteria were not able to grow on liquid 2,2-DCP minimal medium possibly due to the toxicity of the compound to the cell culture or the gene lacking of permease uptake system to transport the substrate into the cells and/or dehalogenase gene is silence. This is the first reported case that the bacteria isolated from the gut of the freshwater fish, Labeo rohita capable of degrading halo-aliphatic compound as sole carbon source. It was hypothesized that these strains might play a significant role in the digestion of specialized substrates which few animals can digest. The presence of silence putative dehalogenase gene in the chromosomal DNA of the bacteria that could not grow in 2,2-DCP is far from clear. In future, this study can help to describe the diversity of the microbial dehalogenases and the dehalogenase enzyme regulation.
5. ACKNOWLEDGEMENTS
Eleanor Abel would like to thank Microbiology and Molecular Laboratory, Universiti Teknologi Malaysia (UTM)/Ministry of Higher Education (MOHE)/GUP under Vot.QJ130000.7135.00H34 and Government of Japan for financial support.
REFERENCES
- Fetzner S. (1998) Bacterial dehalogenation. Applied Microbiology and Biotechnology, 50, 633-657. doi:10.1007/s002530051346
- Magee, L.A. and Colmer, A.R. (1959) Decomposition of 2,2-dichloropropionic acid by soil bacteria. Canadian Journal of Microbiology, 5, 255-260. doi:10.1139/m59-030
- Jing, N.H. and Huyop, F. (2008) Enzymatic dehalogenation of 2,2-dichloropropionic acid by locally isolated Methylobacterium sp. HJ1. Journal of Biological Science, 8, 233-235. doi:10.3923/jbs.2008.233.235
- Olaniran, A.O., Pillay, D. and Pillay, B. (2004) Haloalkane and haloacid dehalogenases from aerobic bacterial isolates indigenous to contaminated sites in Africa demonstrate diverse substrate specificities. Chemosphere, 55, 27-33. doi:10.1016/j.chemosphere.2003.10.067
- Huang, J., Xin, Y., Cao, X. and Zhang, W. (2011) Phylogenetic diversity and characterization of 2-haloacid degrading bacteria from the marine sponge Hymeniacidon perlevis. World Journal of Microbiology and Biotechnology, 27, 1787-1794. doi:10.1007/s11274-010-0636-8
- Hang, J., Xin, Y., Zhang, W. (2011) Isolation, characterization and identification of a Paracoccus sp. 2-haloaciddegrading bacterium from the marine sponge Hymeniacidon perlevis. Journal of Basic Microbiology, 51, 318- 324. doi:10.1002/jobm.201000205
- Ismail, N., Taha, A.M., Jing, N.H., Wahab, R.A., Hamid, A.A.A., Pakingking, R.V. Jr. and Huyop, F. (2008) Biodegradation of monochloroacetic acid by a presumptive Pseudomonas sp. strain R1 bacterium isolated from Malaysian paddy (rice) field. Biotechnology, 7, 481-486. doi:10.3923/biotech.2008.481.486
- Thasif, S., Hamdan, S. and Huyop, F. (2009) Degradation of D,L-2-chloropropionic acid by bacterial dehalogenases that shows stereospecificity and its partial enzymatic characteristics. Biotechnology, 8, 264-269. doi:10.3923/biotech.2009.264.269
- Govender, A., Shaik, R., Nathlee, S.A. and Pillay, B. (2011) Dehalogenase gene detection and microbial diversity of a chlorinated hydrocarbon contaminated site. World Journal of Microbiology and Biotechnology, 27, 2407-2414. doi:10.1007/s11274-011-0713-7
- Chiba, Y., Yoshida, T., Ito, N., Nishimura, H., Imada, C., Yasuda, H. and Sako, Y. (2009) Isolation of a bacterium possessing a haloacid dehalogenase from a marine sediment core. Microbes Environment, 24, 276-279. doi:10.1264/jsme2.ME09123
- Hill, K.E., Marchesi, J.R. and Weightman, A.J. (1999) Investigation of two evolutionarily unrelated halocarboxylic acid dehalogenase gene families. Journal of Bacteriology, 181, 2535-2547.
- Mesri, S., Wahab, R.A. and Huyop, F. (2009) Degradation of 3-chloropropionic acid by Pseudomonas sp. B6P isolated from a rice paddy field. Annals of Microbiology, 59, 447-451. doi:10.1007/BF03175129
- Huyop, F. and Nemati, M. (2010) Properties of dehalogenase in Rhizobium sp. RC1. African Journal of Microbiology Research, 4, 2836-2847.
- Weightman, A.J., Weightman, A.L. and Slater, J.H. (1982) Stereospecificity of 2-monochloropropionate dehalogenation by two dehalogenases of Pseudomonas putida PP3: Evidence for two different dehalogenation mechanisms. Journal of General Microbiology, 128, 1755-1762.
- Huyop, F. and Cooper, R.A. (2011) Regulation of dehalogenase E (DehE) and expression of dehalogenase regulator gene (DehR) from Rhizobium sp. RC1 in E. coli. Biotechnology and Biotechnological Equipment, 25, 2237- 2242. doi:10.5504/bbeq.2011.0009
- Chan, W.Y., Wong, M., Guthrie, J., Savchenko, A.V., Yakunin, A.F., Pai, E.F. and Edwards, E.A. (2010) Sequenceand activity-based screening of microbial genomes for novel dehalogenases. Microbial Biotechnology, 3, 107-120. doi:10.1111/j.1751-7915.2009.00155.x
- Huyop, F. and Sudi, I.Y. (2012) D-specific dehalogenases, a review. Biotechnology and Biotechnological Equipment, 26, 2817-2822. doi:10.5504/bbeq.2011.0143
- Hamid, T.H.T., Hamid, A.A.A. and Huyop, F. (2011) A review: On non-stereospecific haloalkanoic acid dehalogenases. African Journal of Biotechnology, 10, 9725-9736. doi:10.5897/AJB11.934
- Sahoo, M. and Mukhopadhyay, P.K. (2008) Dietary ascorbic acid requirements of fingerlings of genetically improved rohu (Labeo rohita Ham). Israel Journal of AquacultureBamidgeh, 60, 100-106.
- Chebbi, S.G. and David, M. (2010) Respiratory responses and behavioural anomalies of the carp Cyprinus carpio under quinalphos intoxication in sublethal doses. Science Asia, 36, 12-17. doi:10.2306/scienceasia1513-1874.2010.36.012
- James, R. (2010) Effect of dietary supplementation of spirulina on growth and phosphatase activity in copperexposed carp (Labeo rohita). Israel Journal of Aquaculture-Bamidgeh, 62, 19-27.
- Stouthart, X.J.H.X., Haans, J.L.M., Lock, A.C. and Wendelaar Bonga, S.E. (1996) Effects of water pH on copper toxicity to early life stages of the common carp (Cyprinus carpio). Environmental Toxicology and Chemistry, 15, 376- 383. doi:10.1002/etc.5620150323
- Hareland, W.A., Crawford, R.L., Chapman, P.J. and Dagley, S. (1975) Metabolic function and properties of 4-hydroxyphenyl-acetic acid 1 hydroxylase from Pseudomonas acidovorans. Journal of Bacteriology, 121, 272-285.
- Fulton C.K. and Cooper, R.A. (2005) Catabolism of sulfamate by Mycobacterium sp. CF1. Environmental Microbiology, 7, 378-381. doi:10.1111/j.1462-2920.2004.00719.x
- Sambrook, J., Fritsch, E.F. and Maniatis, T. (1989) Molecular cloning: A laboratory manual. 2nd Edition, Cold Spring Harbour Laboratory Press, New York.
- Bergman, J.G. and Sanik, J. (1957) Determination of trace amounts of chlorine in naptha. Analytical Chemistry, 29, 241-243. doi:10.1021/ac60122a018
- Gornall, A.G., Bardawill, C.J. and David, M.M (1949) Determination of serum proteins by means of the biuret reaction. Journal of Biological Chemistry, 177, 751-766.
- Holt, J.G., Krieg, N.R., Sneath, P.H.A., Staley, J.T. and Williams, S.T. (1994) Bergey’s manual of determinative bacteriology. 9th Edition, Williams and Wilkins, Baltimore.
- Nubel, U., Garcia, P.F. and Muyzer, G. (1997) PCR primers to amplify 16S rRNA genes from cyanobacteria. Applied and Environmental Microbiology, 63, 3327- 3332.
- Allison, N. (1981) Bacterial degradation of halogenated aliphatic acids. Ph.D. Thesis, Trent Polytechnic, Nottingham.
- Jing, N.H., Wahab, R.A., Hamdan, S. and Huyop, F. (2010) Cloning and DNA sequence analysis of the haloalkanoic permease uptake gene from Rhizobium sp. RC1. Biotechnology, 9, 319-325. doi:10.3923/biotech.2010.319.325
- Yu, M., Faan, Y.W., Chung, W.Y.K. and Tsang, J.S.H. (2007) Isolation and characterization of a novel haloacid permease from Burkholderia cepacia MBA4. Applied and Environmental Microbiology, 73, 4874-4880. doi:10.1128/AEM.00576-07
- Ghosh, K., Sen, S.K. and Ray, A.K. (2002) Characterization of bacilli isolated from the gut of rohu, Labeo rohita, fingerlings and its significant in digestion. Journal of Applied Aquaculture, 12, 33-42. doi:10.1300/J028v12n03_04
- Bairagi, A., Ghosh, K.S., Sen, S.K. and Ray, A.K. (2002) Enzyme producing bacteria bacterial flora isolated from fish digestive tracts. Aquaculture International, 10, 109- 121. doi:10.1023/A:1021355406412
- Ghosh, K., Roy, M., Kar, N. and Ringo, E. (2010) Gastrointestinal bacteria in Rohu, Labeo rohita (Actinopterygii: Cypriniformes: Cyprinidae), scanning electron microscopy and bacteriological study. Acta Ichthyologica et Piscatoria, 40, 129-135. doi:10.3750/AIP2010.40.2.05
- Kar, N. and Ghosh, K. (2008) Enzyme producing bacteria in the gastrointestinal tracts of Labeo rohita (Hamilton) and Channa punctatus (Bloch). Turkish Journal of Fisheries and Aquatic Sciences, 8, 115-120.
- Ray, A.K., Roy, T., Mondal, S. and Ringo, E. (2010) Identification of gut-associated amylase, cellulose and protease-producing bacteria in three species of Indian major carps. Aquaculture Research, 41, 1462-1469. doi:10.1111/j.1365-2109.2009.02437.x
- Mondal, S., Roy, T., Sen, S.K. and Ray, A.K. (2008) Distribution of enzyme-producing bacteria in the digestive tracts of some freshwater fish. Acta Ichthyologica et Piscatoria, 38, 1-8. doi:10.3750/AIP2008.38.1.01
- Briganti, F., Pessione, E., Giunta, C. and Scozzafava, A. (1997) Purfication, biochemical properties and substrate specificity of a catechol 1,2-dioxygenase from a phenol degrading Acinetobacter radioresistens. FEBS Letters, 416, 61-64. doi:10.1016/S0014-5793(97)01167
- Chibata, I. and Tosa, T. (1981) Use of immobilized cells. Annual Review of Biophysic and Bioengineering, 10, 197- 216. doi:10.1146/annurev.bb.10.060181.001213
- Kafilzadeh, F., Mohammad, S.F. and Yaghoob, T. (2010) Isolation and identification of phenol degrading bacteria from Lake Parishan and their growth kinetic assay. African Journal of Biotechnology, 9, 6721-6726.
- Perpetuo, E.A., Marques, R.C.P., Mendes, M.A., De Lima, W.C., Menck, C.F.M. and Do Nascimento, C.A.O. (2009) Characterization of the phenol monooxygenase gene from Chromobacterium violaceum: Potential use for phenol biodegradation. Biotechnology and Bioprocess Engineering, 14, 694-701. doi:10.1007/s12257-008-0266-2
- Zilli, M., Palazzi, E., Sene, L., Converti, A. and Borghi, M.D. (2001) Toluene and styrene removal from air in biofilters. Process Biochemistry, 10, 423-429. doi:10.1016/S0032-9592(01)00228-X
- Allende, J.L., Gibello, A., Fortun, A., Mengs, G., Ferrer, E. and Martin, M. (2000) 4-hydroxybenzoate uptake in an isolated soil Acinetobacter sp. Current Microbiology, 40, 34-39. doi:10.1007/s002849910007
- Martin, M., Mengs, G., Allende, J.L., Fernandez, J., Alonso, R. and Ferrer, E. (1999) Characterization of two novel propachlor degradation pathways in two species of soil bacteria. Applied and Environmental Microbiology, 65, 802-806.
- Mergeay, M., Monchy, S., Vallaeys, T., Auquier, V., Benotmane, A., Bertin, P., Taghavi, S., Dunn, J., Lelie van der., D. and Wattiez, R. (2003) Ralstonia metallidurans, a bacterium specifically adapted to toxic metals: Towards a catalogue of metal-responsive genes. FEMS Microbiology Reviews, 27, 385-410. doi:10.1016/S0168-6445(03)00045-7
- Song, J.S., Lee, D.H., Lee, K. and Kim, C.K. (2003) Characteristic of several bacterial isolates capable of degrading chloroaliphatic compounds via hydrolytic dechlorination. Journal of Microbiology, 41, 277-283.
- Stringfellow, J.M., Cairns, S.S., Cornish, A. and Cooper, R.A. (1997) Haloalkanoate dehalogenase II (DehE) of a Rhizobium sp. molecular analysis of the gene and formation of carbon monoxide from trihaloacetate by the enzyme. European Journal of Biochemistry, 250, 789-793. doi:10.1111/j.1432-1033.1997.00789.x
- Nardi-Dei, V., Kurihara, T., Park, C., Esaki, N. and Soda, K. (1997) Bacterial D,L-2-haloacid dehalogenase from Pseudomonas sp. strain 113: Gene cloning and structural comparison with D-and L-2-haloacid dehalogenases. Journal of Bacteriology, 179, 4232-4238.
- Topping, A.W. (1992) An investigation into the transposition and dehalogenase functions of DEH, a mobile genetic element, from Pseudomonas putida strain PP3. Ph.D. Thesis, University of Wales, Cardiff.
- Liu, J.Q., Kurihara, T., Hasan, A.K., Nardi-dei, V., Koshikawa, H., Esaki, N. and Soda, K. (1994) Purification and characterization of thermostable and non-thermostable 2- haloacid dehalogenases with different stereospecificities from Pseudomonas sp. strain YL. Applied and Environmental Microbiology, 60, 2389-2393.
- Tsang, J.S.H and Sam, L. (1999) Cloning and characterization of a cryptic haloacid dehalogenase from Burkholderia cepacia MBA4. Journal of Bacteriology, 181, 6003- 6009.
NOTES
*Corresponding author.

