Advances in Microbiology
Vol.2 No.4(2012), Article ID:25847,12 pages DOI:10.4236/aim.2012.24061
Virulence Markers in Staphylococcus aureus Strains Isolated from Hemodialysis Catheters of Mexican Patients
1FES-Iztacala, Universidad Nacional Autónoma de México (UNAM), Tlalnepantla, México
2Ciencias Biológicas y de la Salud, Universidad Autonoma Metropolitana (UAM)—Xochimilco, Calzada del Hueso, México
3Departamento de Sistemas Biológicos, División de Ciencias Biológicas y de la Salud, Universidad Autonoma Metropolitana (UAM)—Xochimilco, Calzada del Hueso, México
4Hospital General Regional 72, Instituto Mexicano del Seguro Social, Gustavo Baz Esquina F. Gómez, Tlalnepantla, México
Email: *vacasergio@gmail.com
Received September 25, 2012; revised November 6, 2012; accepted November 12, 2012
Keywords: S. aureus; Hemodialysis; Catheter; Genotyping
ABSTRACT
Methicillin-resistant Staphylococcus aureus is an increasingly important cause of nosocomial bacterial infections worldwide. S. aureus is responsible for several pathologies, including skin infections, endocarditis, meningitis, deeptissue ulcers, and sepsis. S. aureus biofilm formation on catheters and other medical devices is a major post-operative concern, because biofilms are often the source of persistent and difficult to treat bacterial infections. While catheterrelated S. aureus infections have been reported, the strains responsible for these infections have not been genetically characterized. We genetically characterized S. aureus strains isolated from hemodialysis catheters in Mexican patients. The frequency of 35 genes coding for adhesins, toxins, and other virulence-associated products in the 55 isolated S. aureus strains was determined using PCR, while real-time PCR was used to examine the level of gene expression. Of the 55 S. aureus strains isolated from 109 patients, 45 (81.8%) were determined to be methicillin-resistant. The icaA, rbf, sarA, and agr genes are involved in biofilm formation and bacterial dispersion and were detected in 96.3%, 40.0%, 74.5%, and 100% of S. aureus strains, respectively, and 70.9% of the strains formed a detectable biofilm. Interestingly, 67.2% of the strains contained the icaA, agr, spa, clfA, sdrC, sdrD, sdrE, seg, seh, and sei genes, suggesting that this gene combination is important for successful catheter colonization. The results of this study provide significant insight into the virulence gene make-up of catheter-colonizing S. aureus strains, and will assist in developing a more targeted treatment approach for persistent S. aureus biofilm contamination of medical devices.
1. Introduction
Staphylococcus aureus is a frequent cause of catheterrelated infections in patients [1]. The worldwide emergence of methicillin-resistant S. aureus (MRSA) is of major concern, as its emergence has dramatically reduced the number of antibiotics available for the prevention and treatment of infections in both hospitals and communities [2]. In 2005 in the United States, invasive MRSA infections occurred in 42.5/1000 dialysis population, a rate exceeding that in the general population by 100-fold [3]. MRSA exhibits a great capacity for biofilm formation on surfaces, making endovascular catheters a favorable nidus for infection. S. aureus formation of biofilms requires the synthesis of PNAG (polymeric N-acetylglucosamine), and the enzymes responsible for its synthesis are encoded by the icaADBC operon [4]. Additionally, the rbf gene promotes biofilm formation by S. aureus via repression of icaR, a negative regulator of the icaADBC operon [5]. The SarA protein, which is encoded by the sarA locus, is a positive regulator of PNAG-dependent S. aureus biofilm formation [6,7].
The microbial adherence to cells and extracellular matrix is considered as an essential first step in the process of colonization and infection [8]. A well characterized family of staphylococcal surface adhesins, called MSCRAMMs (microbial surface components recognizing adhesive matrix molecules) are known to mediate adherence to host extracellular matrix components, such as fibrinogen, fibronectin and collagen [9].
Among the S. aureus adhesin-coding genes, fnbA, fnbB, spa, clfA, clfB, cna, bbp, ebps, map/eap, sdrC, sdrD and sdrE are found (see Table 1). S. aureus strains harboring fnbA and fnbB have been found associated with invasive infections such as endocarditis, septic ar-
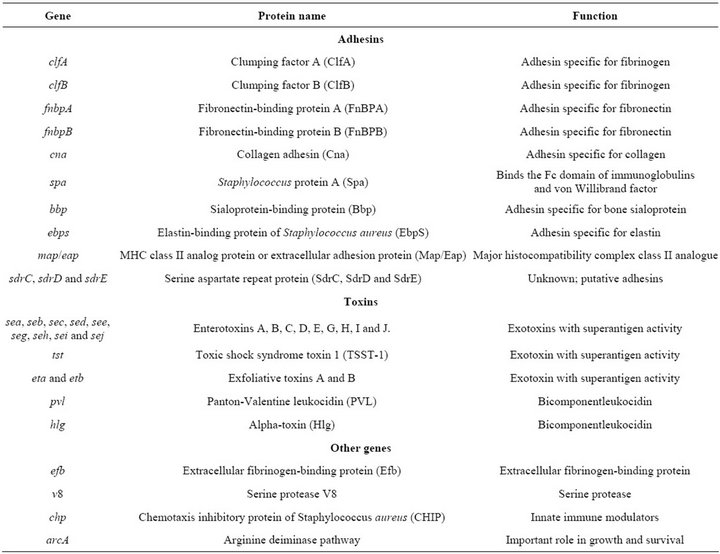
Table 1. Virulence-related genes studied in the S. aureus strains [25].
thritis and osteomyelitis [10]. It has also been shown that protein A (Spa) is an important virulence factor in septic arthritis models and subcutaneous infections in mice [11]. S. aureus strains deficient in CNA showed impaired capacity to produce endocarditis [12], osteomyelitis [13] and keratitis [14]. It has been shown that clumping factor A (ClfA) is an important S. aureus virulence factor in several experimental infection models, including rat endocarditis [15], mice arthritis [16] and rabbit infective endocarditis [17]. ClfA also inhibits phagocytosis by human polymorphonuclear leucocytes [18].
S. aureus is able to produce a great number of extracellular proteins, including enterotoxins (SEA, SEB, SEC, SED, SEE, SEG, SEH, SEI and SEJ), PantonValentine leukocidin (PVL), toxic shock syndrome toxin 1 (TSST-1), exfoliative toxin (ETA and ETB) and alpha toxin (Hlg), some of which can act as superantigens (Table 1).
Other virulence factors are encoded by the genes chp, v8, arcA and coa [19]. The expression of most of the virulence factors in S. aureus is controlled by the agr locus [20].
Despite the fact that severe catheter-related S. aureus infections have been reported [21-23], and that these infections have become important causes of morbidity, mortality, and a source of concern in the primary and emergency care context over the past decade [24], the S. aureus strains responsible for these infections have not been genetically characterized. Therefore, in order to gain some insight into the combinations of genes present in catheter-colonizing S. aureus strains, the presence of 35 virulence genes coding for adhesins, toxins, or other products was determined in 55 S. aureus strains isolated from tunneled hemodialysis catheters of Mexican patients with chronic renal failure being treated with hemodialysis. Hence we screened by PCR for the presence of 35 common S. aureus virulence-associated genes, including adhesin, toxin, superantigen, and biofilm formation genes. We then used real-time PCR to measure the expression of icaA, rbf, and sarA biofilm formation genes, as well as that of the global virulence gene regulator, agr. This study is the first report on the prevalence of the 35 most common virulence-associated genes in S. aureus strains isolated from hemodialysis catheters. These findings will be of significant interest to health professionals and researchers alike, as the discovery of gene sets required for catheter colonization will allow for a more targeted approach to the development of treatment strategies, which would be of great interest to vaccine researchers.
2. Materials and Methods
2.1. Patients
A total of 109 patients presenting with symptoms of infection (e.g., erythema, inflammation, pain, and yellow suppuration) at the insertion site of a Mahurkar-type catheter were selected during September 2009 to May 2010. All of the patients had visited hemodialysis services at four public hospitals located in Estado de Mexico, Mexico [40.4% (n = 44) of the samples were from Hospital General Regional 72 del IMSS; 26.6% (n = 29) from Hospital General Regional N˚ 196 Aragón; 21.1% (n = 23) from ISSEMyM Satélite; and 11.9% (n = 13) of the samples were from Unidad Médica de Atención Ambulatoria (UMAA) N˚ 199]. The patients ranged in age from 17 - 77 years old, with 56% (n = 61) of the patients less than 50 years old. The causes of the chronic renal insufficiency were diabetes mellitus (33% n = 36), glomerulonephritis (17.4%, n = 19), hypertension (10.1%, n = 11), polycystic kidney disease (1.84%, n = 2), renal failure secondary to neurogenic bladder, and congenital malformation, lupus nephritis, or preeclampsia (each with 0.92%, n = 1); the cause was not determined in 34% of the patients (n = 37). The local ethics committee of each hospital approved the study. Inclusion criteria for choosing patients were: persons suffering chronic renal failure, with symptoms of infection and accepting to participate in the study by sign in the informed consent letter. Patients who were under treatment with antibiotics at the moment of sampling or during the last 30 days were excluded. Patients not accepting participate in the study were also excluded.
2.2. Sampling, Bacteria Identification, and PCR Conditions
After obtaining informed consent from each patient, samples were taken from the exterior terminal end of the catheter using sterile cotton swabs. The samples were placed in Stuart transport medium (BD Bioxon, Cuautitlán Izcalli, Edo. de Mexico, Mexico) and taken to the laboratory for further cultivation. For the primary phenotypical species identification of the S. aureus strains, tube-coagulase test (Bactident-coagulase, Merck, Darmstadt, Germany) and Api 32 Staph system (BioMerieux, Durham, NC, USA) were applied. This was confirmed by molecular detection of the S. aureus specific genes encoding 23S rRNA, thermostable nuclease (nuc), clumping factor (clfA), coagulase (coa) and protein A region X (spa), and the femA and femB genes. Table 1 shows the virulence-related genes studied in the S. aureus strains. Bacterial DNA was extracted using a Wizard Genomic DNA Purification Kit (Promega, Madison, WI). One ml of an overnight culture was centrifuged at 13,000 rpm for 2 minutes and the pelleted cells were resuspended in 480 µL of 50 mM EDTA. A mixture of 60 µL of 10 mg/mL lysozyme (Sigma Cat. # L7651) and 60 µL of 10 mg/mL lysostaphin (Sigma Cat. # L7386) were added and the sample was incubated at 37˚C for 60 minutes. Afterwards sample was centrifuged 2 minutes at 13,000 rpm and the supernatant was removed. Six hundred microliters of Nuclei Lysis solution were added in order to resuspend the cells. Sample was incubated at 80˚C for 5 minutes to lyse the cells and then cooled to room temperature. Three microliters of RNase solution were added and the tube was inverted 5 times to mix. Sample was incubated at 37˚C for 60 minutes and then cooled to room temperature; 200 µL of Protein Precipitation Solution were added to the RNase-treated cell lysateand the mixture was vortexed vigorously at high speed for 20 seconds. Sample was incubated on ice for 5 minutes, centrifuged at 13,000 rpm for 3 minutes, and the supernatant containing the DNA was transferred to a clean 1.5 mL microcentrifuge tube containing 600 µL of room temperature isopropanol. The tube was mixed by inversion and centrifuged at 13,000 rpm for 2 minutes. Supernatant was poured off and the tube was drained on absorbent paper. Afterwards, 600 µL of room temperature 70% ethanol were added and the tube was gently inverted several times to wash the DNA pellet. The tube was centrifuged at 13,000 rpm for 2 minutes and the ethanol was aspirated. The tube was drained on absorbent paper and the pellet was allowed to air-dry for 15 minutes. One hundred microliters of DNA rehydration solution were added to the tube and the sample was incubated at 65˚C for 1 hour. DNA was stored at –20˚C until it was used.
PCR amplification of genetic markers was performed in a Corbette Research Thermocycler using PuReTaq Ready-To-Go PCR Beads (GE Healthcare, Piscataway, New Jersey, USA). The amplified products were stained with ethidium bromide after electrophoresis in a 1% or 2% agarose gel. PCR primers, primers and probes for real time PCR and conditions used in PCR and real time PCR assays are listed in Table 2. The ATCC 33592 S. aureus strain was used as a positive control for PCR amplification of fnbA, fnbB, spa, clfA, clfB, cna, ebps, map/eap, sdrC, sdrD, sdrE, icaA, rbf, sarA and group I agr loci. For bbp, eta, etb, hlg, pvl, sea, seb, sec, sed, see, seg, seh, sei, sej, tst, chp, efb, V8, arcA and coa, the following strains from our collection were used, respec-

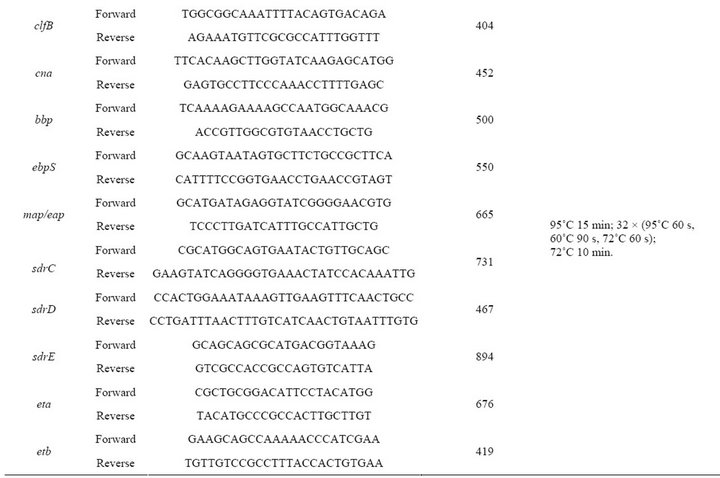


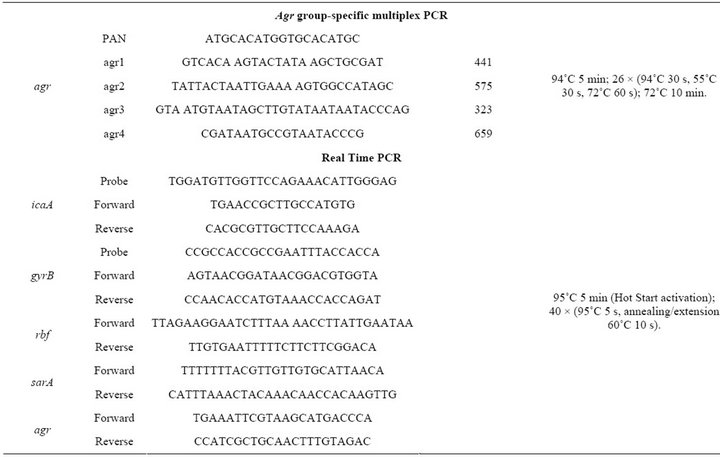
Table 2. PCR primers, primers and probes for real time PCR and conditions used in PCR and real time PCR assays.
tively: sa93, sa19, sa55, sa75, sa59, sa9, sa32, sa79, sa66, sa73, sa31, sa33, sa62, sa15, sa63, sa110, sa111, sa43, sa105 and sa35.
The expression of gyrB, icaA, rbf, sarA and agrA was measured using RT-PCR. To 500 μL of bacterial culture, grown in TSB at 37˚C for 24 h, 1000 μL of RNA Protect Bacteria Reagent (Qiagen, Hilden, Germany) were added. Sample was vortexed for 30 seconds and incubated at room temperature for 5 minutes. Sample was centrifuged at 9400 rpm for 10 minutes and the bacterial cells were resuspended in 200 μL of TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8) containing 10 mg/mL lysozyme and 40 mg/mL lysostaphin. Sample was vortexed by 10 seconds and incubated at room temperature for 5 minutes. Afterwards, extraction and purification of total RNA was done using the RNeasy Mini Kit (Qiagen, Hilden, Germany), according to the manufacturer’s instructions, including on-column DNase treatment. The concentration and purity of total RNA were analyzed using a NanoDrop 2000 spectrophotometer (NanoDrop Technologies, Inc.).
To obtain cDNA, a QuantiTec Reverse transcription kit (Qiagen), which includes genomic DNA elimination, was used according to the manufacturer’s instructions. The icaA probe contained the fluorescent reporter dye FAM covalently linked to the 5’end and BHQ-1 covalently linked to the 3’end as a quencher. The gyrB probe contained the fluorescent reporter dye JOE covalently linked to the 5’end and BHQ-1 covalently linked to the 3’end as a quencher. A Rotor Gene Probe PCR Kit (Qiagen, Hilden, Germany) and Rotor Gene SYBR Green PCR Kit (Qiagen) kit were used for the real-time PCR assays.
The sa22, sa103 S. aureus clinical strains from our collection, S. epidermidis ATCC 35984 and E. coli ATCC 11775 were used as negative controls for the realtime PCR assays. S. aureus ATCC 33592 was used as the positive control.
2.3. Determination of Biofilm Formation
S. aureus biofilm formation was determined using the microtiter plate assay, as described in [15]. S. epidermidis ATCC 35984 and S. epidermidis ATCC 12228 were used as positive and negative controls, respectively. In this assay, polystyrene-adhered bacteria were stained with safranin, and the absorbance at 492 nm was recorded. Strains with an A492 less than 0.2, between 0.2 and 1.0, or greater than 1.0 were considered biofilm negative, weak biofilm formers, or strong biofilm formers, respectively [26].
2.4. Statistical Analysis
The frequencies of the virulence-related gene combinations among the MRSA (mecA+) and MSSA (mecA−) S. aureus strains were analyzed using Chi-squared tests. A p value less than 0.005 was considered significant.
3. Results
Half of the catheter-isolated bacteria (55/109) were identified as S. aureus based on PCR amplification of the 23S rRNA, nuc, clfA, coa, spa, femA and femB genes. The remaining isolates (n = 54) were identified as S. epidermidis.
The most frequently detected virulence genes in the analyzed strains were i) adhesin-coding genes, including spa (100%), clfA (100%), sdrD (94.5%), sdrE (92.7%), sdrC (89%) and ebps (85.4%); ii) toxin-coding genes, including hlg (96.3%), seg (94.5%), sei (94.5%) and seh (78.1%); and iii) genes implicated in biofilm formation, including icaA (96.3%), sarA (74.5%) and rbf (40%) (Table 3). The agr operon was detected in all S. aureus strains; 16 strains harbored the group I agr locus, 29 the group II agr locus and 10 the group III agr locus (Table 3). Almost 82% of the strains were mecA+. None of the strains harbored the pvl or sej genes (Table 3).
The strains contained combinations of 15 to 25 of the 35 examined virulence genes (Table 4). Six strains (11%) carried a combination of 25 genes, with 12 genes coding for adhesins with affinity for fibronectin, fibrinogen and collagen and 5 genes coding for toxins, some of which can act as superantigens (Tables 1 and 4). Sixty percent of the strains carried a combination of the seg, seh and sei toxin genes (Table 4).
Fifteen strains carried six (fnbA, cna, sdrE, eta, hlg, ica) of the seven virulence genes that have been reported to be significantly more common in invasive S. aureus strains [19]. Six of these fifteen strains carried the 25- gene combination (Table 4), and the remaining nine strains each harbored a unique combination of virulence genes (data not shown).
3.1. Detection of IcaA, Rbf, SarA and AgrA Expression Using Real Time PCR
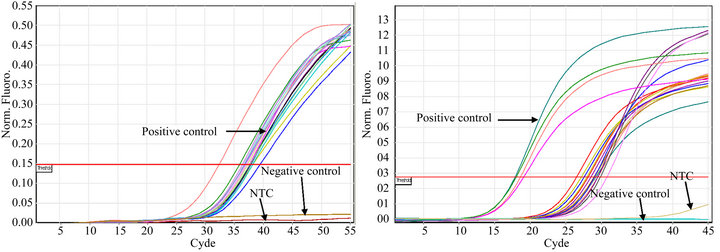
All of the S. aureus strains carrying icaA (n = 53), rfb (n = 22), sarA (n = 41) or agr (n = 55) expressed the genes (Figures 1(a)-(d)). Constitutive gyrB expression was used as a control (data not shown). The Tm was determined for each real time PCR run, when Rotor Gene SYBR Green PCR Kit was used, to differentiate specific amplification from nonspecific amplification (data not shown).
3.2. Determination of Biofilm Formation
Among the S. aureus strains studied, 70.9% (n = 39) were able to form a biofilm on polystyrene; 51% (n = 28) were weak biofilm formers, and 20% (n = 11) were strong biofilm formers.

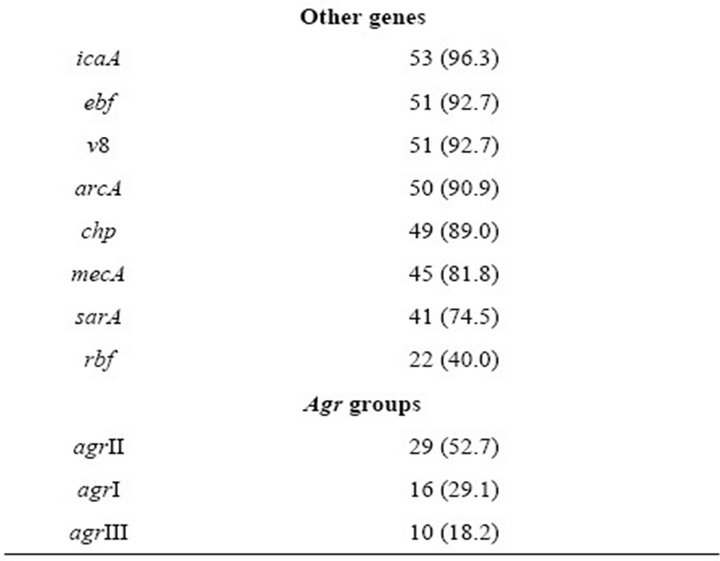
Table 3. Frequency of mecA and virulence-related genes in S. aureus strains.
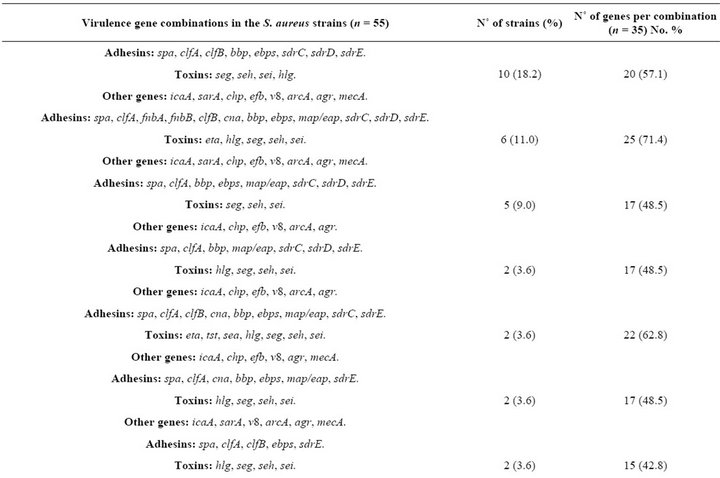
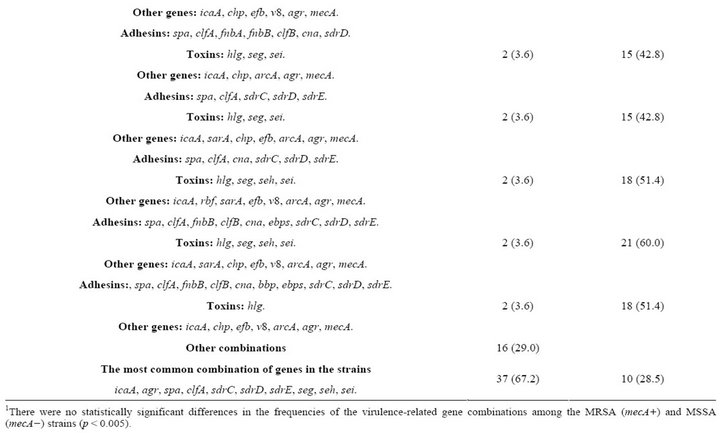
Table 4. Combinations of virulence-related genes in S. aureus strains1.
 (a)
(a)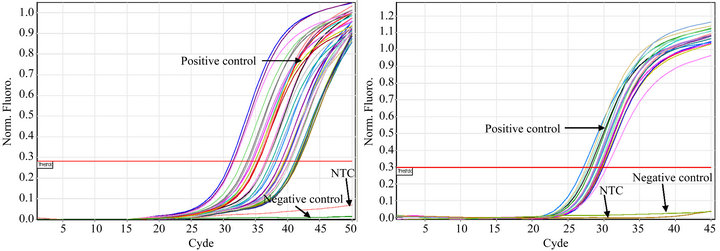 (b)
(b)
Figure 1. Detection of icaA (a), rbf (b), sarA (c), and agrA (d) expression in S. aureus strains isolated from catheters using real time PCR. S. aureus ATCC 33592 was used as a positive control; the negative controls were S. epidermidis ATCC 35984 (a), sa22 (b), sa103 (c) and E. coli ATCC 11775 (d). NTC indicates the no DNA control.
4. Discussion
In this study, we found that 55 of 109 catheters in hemodialysis patients harbored S. aureus. This high incidence of S. aureus-positive catheters may be due to a deficiency in the management of patients within the different hospital settings and/or the high rate of autoinfection by the patients’ own indigenous S. aureus, which is present in their anterior nares [27].
Despite reports of severe catheter-related S. aureus infections [21-23,28], the S. aureus strains responsible for these infections have not been genetically characterized. While the genotype of S. aureus has been suggested to contribute to the severity of the infection [29], the S. aureus virulence factors that are involved in catheter colonization and the subsequent bacteremia are not known. Bacterial adhesins, toxins, and genes involved in bacterial dispersion may be involved in the development of catheter-related infections. Therefore, to gain some insight into the combinations of genes present in cathetercolonizing S. aureus strains, the presence of 35 virulence genes coding for adhesins, toxins, or other products was determined in S. aureus strains isolated from tunneled hemodialysis catheters. As expected, a large number of strains carried genes for at least five adhesins (maximum of 12, average of 7; Table 4), with the combination of spa, clfA, sdrC, sdrD and sdrE being the most frequently detected combination. The Sdr proteins in S. aureus are members of the MSCRAMMs family (Microbial Surface Components-Recognizing Adhesive Matrix Molecules) that are encoded by the tandemly arrayed sdrC, sdrD and sdrE genes [30]. Although the precise role of Sdr adhesins in staphylococcal infection is not known, a strong correlation between the sdr genes of S. aureus and certain diseases has been reported. There is a significantly increased prevalence of the sdrE gene in invasive S. aureus strains [31], in S. aureus strains responsible for osteomyelitis [32], and in S. aureus isolates responsible for bone infections [33].
Among the toxin genes, hlg (96.3%), seg (92.7%), sei (85.4%) and seh (78.1%) were the most frequently detected; no sej-positive or pvl-positive strains were found, and the most frequently observed combination was seg, seh, and sei (74.5%; Table 4). The frequency of toxinencoding genes in the catheter-colonizing strains was higher than the previously reported frequency in a series of 100 MRSA isolates from hospital infections; with seg (77%) and sei (77%) being the most frequently detected toxin-encoding genes [34]. However, it has been reported that invasive S. aureus strains were not more likely than noninvasive strains to carry the seg, seh and sei genes [31].
The virulence markers ebf, v8, arcA and chp were identified in most S. aureus strains (Table 2), with frequencies similar to those reported for S. aureus strains isolated from complicated skin and skin-structure infections [35].
All strains carrying genes involved in biofilm formation and bacterial dispersion expressed the genes (Figure 1), and 70.9% of the examined strains formed a biofilm. This is an important bacterial virulence property as it has been reported that staphylococci account for more than 60% of all identified pathogens in central venous catheter-related infections [36]. Moreover, in a Center for Disease Control report of 2005, 15% of all reported MRSA infections occurred in dialysis patients [3], and a rate of 0.88 MRSA bacteremias/100 hemodialysis patients/year was reported in England for 2008-2009 [37].
Thirty-seven S. aureus isolates (67.2%) had the virulence gene combination of icA, agr, spa, clfA, sdrC, sdrD, sdrE, seg, seh and sei (Table 4), a combination that was detected in 16 different gene patterns (Table 4); 30 of these strains were mecA+. This result suggests that this gene combination may be important for successful catheter colonization. We conclude that these S. aureus strains containing such array of virulence factors are an element of risk for catheterized Mexican patients, which may acquire bacteremia or other pathologies.
These findings could be used to develop more targeted treatment strategies for S. aureus infections, which would be of great interest to vaccine researchers. The discovered gene combination could also be used to quickly identify S. aureus strains that are a greater threat to catheterized patients, which would warrant more aggressive treatment in a hospital setting.
5. Acknowledgements
This work was supported by grants from the Universidad Nacional Autónoma de México (PAPIIT IN218211) and FES-Iztacala, UNAM (PAPCA 2010-11 Project 7). We would like to thank the Programa de doctorado en Ciencias Biológicas y de la Salud, UAM-Xochimilco, México.
REFERENCES
- P. Viale and S. Stefani, “Vascular Catheter-Associated Infections: A Microbiological and Therapeutic Update,” Journal of Chemotherapy, Vol. 18, No. 3, 2006, pp. 235- 249.
- I. M. Gould, “The Clinical Significance of MethicillinResistant Staphylococcus aureus,” The Journal of Hospital Infection, Vol. 61, No. 4, 2005, pp. 277-282. doi:10.1016/j.jhin.2005.06.014
- Centers for Diseases Control, “Invasive Methicillin-Resistant Staphylococcus aureus Infections among Dialysis Patients—United States, 2005,” Morbidity and Mortality Weekly Report, Vol. 56, No. 9, 2007, pp. 197-199.
- F. Götz, “Staphylococcus and Biofilms,” Molecular Microbiology, Vol. 43, No. 6, 2002, pp. 1367-1378. doi:10.1046/j.1365-2958.2002.02827.x
- D. Cue, M. G. Lei, T. T. Luong, L. Kuechenmeister, P. M. Dunman, S. O’Donnell, S. Rowe, J. P. O’Gara and C. Y. Lee, “Rbf Promotes Biofilm Formation by Staphylococcus aureus via Repression of icaR, a Negative Regulator of icaADBC,” Journal of Bacteriology, Vol. 191, No. 20, 2009, pp. 6363-6373. doi:10.1128/JB.00913-09
- K. E. Beenken, J. S. Blevins and M. S. Smeltzer, “Mutation of SarA in Staphylococcus aureus Limits Biofilm Formation,” Infection and Immunity, Vol. 71, No. 7, 2003, pp. 4206-4211. doi:10.1128/IAI.71.7.4206-4211.2003
- J. Valle, A. Toledo-Arana, C. Berasain, J. M. Ghigo, B. Amorena, J. R. Penadés and I. Lasa, “SarA and Not SigmaB Is Essential for Biofilm Development by Staphylococcus aureus,” Molecular Microbiology, Vol. 48, No. 4, 2003, pp. 1075-1087. doi:10.1046/j.1365-2958.2003.03493.x
- E. Mongodin, O. Bajolet, J. Cutrona, N. Bonnet, F. Dupuit, E. Puchelle and S. de Bentzmann, “FibronectinBinding Proteins of Staphylococcus aureus Are Involved in Adherence to Human Airway Epithelium,” Infection and Immunity, Vol. 70, No. 2, 2002, pp. 620-630.
- W. Navarre and O. Schneewind, “Surface Proteins of Gram-Positive Bacteria and Mechanisms of Their Targeting to the Cell Wall Envelope,” Microbiology and Molecular Biology Reviews, Vol. 63, No. 1, 1999, pp. 174-229.
- S. J. Peacock, N. P. J. Day, M. G. Thomas, A. R. Berendt and T. J. Foster, “Clinical Isolates of Staphylococcus aureus Exhibit Diversity in fnb Genes and Adhesion to Human Fibronectin,” The Journal of Infection, Vol. 41, No. 1, 2000, pp. 23-31. doi:10.1053/jinf.2000.0657
- N. Palmqvist, T. Foster, A. Tarkowski and E. Josefsson, “Protein A Is a Virulence Factor in Staphylococcus aureus Arthritis and Septic Death,” Microbial Pathogenesis, Vol. 33, No. 5, 2003, pp. 239-249. doi:10.1006/mpat.2002.0533
- S. A. Hienz, T. Schennings, A. Heimdahl and J. L. Flock, “Collagen Binding of Staphylococcus aureus Is a Virulence Factor in Experimental Endocarditis,” The Journal of Infectious Diseases, Vol. 174, No. 1, 1996, pp. 83-88. doi:10.1093/infdis/174.1.83
- M. O. Elasri, J. R. Thomas and R. A. Skinner, “Staphylococcus aureus Collagen Adhesin CONTRIBUTES to the Pathogenesis of Osteomyelitis,” Bone, Vol. 30, No. 1, 2002, pp. 275-280. doi:10.1016/S8756-3282(01)00632-9
- M. N. Rhem, E. M. Lech, J. M. Patti, D. McDevitt, M. Höök, D. B. Jones and K. R. Wilhelmus, “The CollagenBinding Adhesin Is a Virulence Factor in Staphylococcus aureus Keratitis,” Infection and Immunity, Vol. 68, No. 6, 2000, pp. 3776-3779. doi:10.1128/IAI.68.6.3776-3779.2000
- P. Moreillon, J. M. Entenza, P. Francioli, D. McDevitt, T. J. Foster, P. Francois and P. Vaudaux, “Role of Staphylococcus aureus Coagulase and Clumping Factor in Pathogenesis of Experimental Endocarditis,” Infection and Immunity, Vol. 63, No. 12, 1995, pp. 4738-4743.
- E. Josefsson, O. Hartford, L. O’Brien, J. M. Patti and T. Foster, “Protection against Experimental Staphylococcus aureus Arthritis by Vaccination with Clumping Factor A, a Novel Virulence Determinant,” The Journal of Infectious Diseases, Vol. 184, No. 12, 2001, pp. 1572-1580. doi:10.1086/324430
- J. Vernachio, A. S. Bayer, T. Le, Y. L. Chai, B. Prater, A. Schneider, B. Ames, P. Syribeys, J. Robbins and J. M. Patti, “Anti-Clumping Factor A Immunoglobulin Reduces the Duration of Methicillinresistant Staphylococcus aureus Bacteremia in an Experimental Model of Infective Endocarditis,” Antimicrobial Agents and Chemotherapy, Vol. 47, No. 11, 2003, pp. 3400-3406. doi:10.1128/AAC.47.11.3400-3406.2003
- J. Higgings, A. Louhhman, K. P. van Kessel, J. A. van Strijp and T. J. Foster, “Clumping Factor A of Staphylococcus aureus Inhibits Phagocytosi by Human Polymorphonuclear Leucocytes,” FEMS Microbiology Letters, Vol. 258, No. 2, 2006, pp. 290-296.
- S. Watanabe, T. Ito, T. Sasaki, S. Li, I. Uchiyama, K. Kishii, K. Kikuchi, R. L. Skov and K. Hiramatsu, “Genetic Diversity of Staphylocoagulase Genes (coa): Insight into the Evolution of Variable Chromosomal Virulence Factors in Staphylococcus aureus,” PLoS One, Vol. 4, No. 5, 2009, Article ID: e5714. doi:10.1371/journal.pone.0005714
- G. Ji, R. C. Beavis and R. P. Novick, “Cell Density Control of Staphylococcal Virulence Mediated by an Octapeptide Pheromone,” Proceedings of the National Academy of Science USA, Vol. 92, No. 26, 1995, pp. 12055- 12059. doi:10.1073/pnas.92.26.12055
- J. J. Ayala-Gaytán, M. C. Alemán-Bocanegra, C. E. Guajardo-Lara and S. B. Valdovinos-Chávez, “Catheter-Associated Bloodstream Infections. Review of Five-Year Surveillance among Hospitalized Patients,” Revista Medica del Instituto Mexicano del Seguro Social, Vol. 48, No. 2, 2010, pp. 145-150.
- G. Jean, “Incidence and Risk Factors for Infections from Hemodialysis Catheters,” Néphrologie, Vol. 22, No. 8, 2001, pp. 443-448.
- G. Jean, B. Charra, C. Chazot, T. Vanel, J. C. Terrat, J. M. Hurot and G. Laurent, “Risk Factors Analysis for LongTerm Tunneled Dialysis-Catheter Related Bacteremia,” Nephron, Vol. 91, No. 3, 2002, pp. 399-405. doi:10.1159/000064279
- M. G. Parker and B. N. Doebbeling, “The Challenge of Methicillin-Resistant Staphylococcus aureus Prevention in Hemodialysis Therapy,” Seminars in Dialysis, Vol. 25, No. 1, 2012, pp. 42-49. doi:10.1111/j.1525-139X.2011.00999.x
- K. Plata, A. E. Rosato and G. Wegrzyn, “Staphylococus aureus as an Infectious Agent: Overview of Biochemistry and Molecular Genetics of Its Pathogenicity,” Acta Biochimica Polonica, Vol. 56, No. 4, 2009, pp. 597-612.
- T. Møretrø, L. Hermansen, A. L. Holck, M. S. Sidhu, K. Rudi and S. Langsrud, “Biofilm Formation and the Presence of the Intercellular Adhesion Locus ica among Staphylococci from Food and Food Processing Environments,” Applied and Environmental Microbiology, Vol. 69, No. 9, 2003, pp. 5648-5655. doi:10.1128/AEM.69.9.5648-5655.2003
- K. Toshkova, C. Annemüller, O. Akineden and C. Lämmler, “The Significance of Nasal Carriage of Staphylococcus aureus as Risk Factor for Human Skin Infections,” FEMS Microbiology Letters, Vol. 202, No. 1, 2001, pp. 17-24. doi:10.1111/j.1574-6968.2001.tb10774.x
- R. Stolić, G. Trajković, V. Perić, A. Jovanović, D. Stolić, S. Sovtić, T. Lazarević, Z. Zivić and G. Subarić-Gorgieva, “Central Venous Catheters in Hemodialysis: To Accept Recommendations or to Stick to Own Experience,” Vojnosanitetski Pregled, Vol. 65, No. 1, 2008, pp. 21-26. doi:10.2298/VSP0801021S
- V. G. Fowler Jr., C. L. Nelson, L. M. McIntyre, B. N. Kreiswirth, A. Monk, G. L. Archer, J. Federspiel, S. Naidich, B. Remortel, T. Rude, P. Brown, L. B. Reller, G. R. Corey and S. R. Gill, “Potential Associations between Hematogenous Complications and Bacterial Genotype in Staphylococcus aureus Infection,” The Journal of Infectious Diseases, Vol. 196, No. 5, 2007, pp. 738-747. doi:10.1086/520088
- E. Josefsson, K. W. McCrea, D. Ni Eidhin, D. O’Connell, J. Cox, M. Höök and T. J. Foster, “Three New Members of the Serine-Aspartate Repeat Protein Multigene Family of Staphylococcus aureus,” Microbiology, Vol. 144, No. 12, 1998, pp. 3387-3395. doi:10.1099/00221287-144-12-3387
- S. J. Peacock, C. E. Moore, A. Justice, M. Kantzanou, L. Story, K. Mackie, G. O’Neill and N. P. Day, “Virulent Combinations of Adhesin and Toxin Genes in Natural Populations of Staphylococcus aureus,” Infection and Immunity, Vol. 70, No. 9, 2002, pp. 4987-4996. doi:10.1128/IAI.70.9.4987-4996.2002
- A. Sabath, D. C. Melles, G. Martirosian, H. Grundmann, A. van Belkum and W. Hryniewicz, “Distribution of the Serine-Aspartate Repeat Protein-Encoding sdr Genes among Nasal and Invasive Staphylococcus aureus Strains,” Journal of Clinical Microbiology, Vol. 44, No. 3, 2006, pp. 1135-1138. doi:10.1128/JCM.44.3.1135-1138.2006
- S. Trad, J. Allignet, L. Frangeul, M. Davi, M. Vergassola, E. Couve, A. Morvan, A. Kechrid, C. Buchrieser, P. Glaser and N. El-Solh, “DNA Macroarray for Identification and Typing of Staphylococcus aureus Isolates,” Journal of Clinical Microbiology, Vol. 42, No. 5, 2004, pp. 2054-2064. doi:10.1128/JCM.42.5.2054-2064.2004
- P. Sauer, J. Síla, T. Stosová, R. Vecerová, P. Hejnar, I. Vágnerová, M. Kolár, V. Raclavsky, J. Petrzelová, Y. Lovecková and D. Koukalová, “Prevalence of Genes Encoding Extracellular Virulence Factors among MeticillinResistant Staphylococcus aureus Isolates from the University Hospital, Olomouc, Czech Republic,” Journal of Medical Microbiology, Vol. 57, No. 4, 2008, pp. 403-410. doi:10.1099/jmm.0.47413-0
- S. J. Campbell, H. S. Deshmukh, C. L. Nelson, I. G. Bae, M. E. Stryjewski, J. J. Federspiel, G. T. Tonthat, T. H. Rude, S. L. Barriere, R. Corey and V. G. Fowler Jr., “Genotypic Characteristics of Staphylococcus aureus Isolates from a Multinational Trial of Complicated Skin and Skin Structure Infections,” Journal of Clinical Microbiology, Vol. 46, No. 2, 2008, pp. 678-684. doi:10.1128/JCM.01822-07
- J. S. Groeger, A. B. Lucas, H. T. Thaler, H. FriedlanderKlar, A. E. Brown, T. E. Kiehn and D. Armstrong, “Infectious Morbidity Associated with Long-Term Use of Venous Access Devices in Patients with Cancer,” Annals of Internal Medicine, Vol. 119, No. 12, 1993, pp. 1168- 1174.
- R. Fluck, J. Wilson and C. R. Tomson, “UK Renal Registry 12th Annual Report (December 2009): Chapter 12: Epidemiology of Methicillin Resistant Staphylococcus aureus Bacteraemia amongst Patients Receiving Dialysis for Established Renal Failure in England in 2008: A Joint Report from the UK Renal Registry and the Health Protection Agency,” Nephron Clinical Practice, Vol. 115, No. 1, 2010, pp. 261-270. doi:10.1159/000301235
NOTES
*Corresponding author.

