World Journal of Cardiovascular Diseases
Vol.07 No.04(2017), Article ID:75345,14 pages
10.4236/wjcd.2017.74010
Integrated Assessment of Cardiac PET/MRI: Co-Registered PET and MRI Polar Plots by Mutual MR-Based Segmentation of the Left Ventricular Myocardium
Felix Nensa1*, Thorsten D. Poeppel2, Ercan Tezgah3, Philipp Heusch4, Kai Nassenstein1, Michael Forsting1, Andreas Bockisch2, Raimund Erbel3, Thomas Schlosser1
1Department of Diagnostic and Interventional Radiology and Neuroradiology, University Hospital Essen, University of Duisburg-Essen, Essen, Germany
2Clinic for Nuclear Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany
3Clinic for Cardiology, University Hospital Essen, University of Duisburg-Essen, Essen, Germany
4Department of Diagnostic and Interventional Radiology, University Hospital Dusseldorf, University of Dusseldorf, Dusseldorf, Germany

Copyright © 2017 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY 4.0).
http://creativecommons.org/licenses/by/4.0/



Received: December 23, 2016; Accepted: April 8, 2017; Published: April 11, 2017
ABSTRACT
Background: In the present study, we sought to describe a procedure for the creation of co-registered positron emission tomography (PET) and magnetic resonance imaging (MRI) polar plots of cardiac PET/MRI examinations, validate the resulting plots against available standard methods in patients with myocardial infarction and provide examples that demonstrate the advantage of the novel approach over existing standards. Methods: Co-registered LGE and PET short-axis images were transformed into polar maps based on a radial sampling pattern. LGE was automatically detected using an automated thresholding algorithm (ATA). In 20 PET/MRI examinations in patients with acute myocardial infarction, agreement between manual LGE assessment and the ATA classification was calculated. Also agreement between MRI-seg- mentation based PET polar plots and standard PET polar plots (created with the Corridor4DM software package) was assessed. Results: No statistically significant difference in infarct sizes between manual and ATA segmentation was found (p = 0.12). Both methods were highly correlated (Pearson’s r = 0.96, p < 0.01). Bland-Altman analysis revealed lower and upper limits of agreement of −4.7 g and 3.6 g. Agreement between MRI-segmentation based PET polar plots and standard PET polar plots was very high (mean kappa of 0.91 ± 0.10; p < 0.01 in all cases). In three additional patients with myocardial inflammation, the software successfully created polar plots that demonstrate the location and extent of pathologic tracer uptake in the left ventricular myo- cardium. Conclusion: A straightforward software approach for the creation of co-registered PET and MRI polar plots was described and successfully de- monstrated in PET/MRI studies of myocardial infarction and inflammation.
Keywords:
Cardiac PET/MRI, Bull’s Eye, Polar Plot, Polar Map

1. Introduction
Since the availability of integrated positron emission tomography (PET)/mag- netic resonance imaging (MRI) devices the simultaneous assessment of cardiac PET and MRI may be performed [1] [2] [3] [4] . However, for now, there is a lack of software tools that allow for an integrated reading of both modalities. Several software packages for the reading of cardiac PET or cardiac MRI exist, but these are dedicated to the use with either modality alone, and thus cannot take advantage of the spatial and temporal coherence of simultaneous image acquisition. Apart from that, numerous packages with generic image fusion capabilities are available. However, these do not incorporate the typical workflows for the reading of cardiac imaging studies. In particular, the comparison of left ventricular MRI and PET findings regarding their anatomical location and extent turned out to be impractical using available standard software [1] . Based on our experience and needs in clinical and scientific reading of cardiac PET/MRI studies, we developed a straightforward and robust software approach for the creation of co-registered parametric polar plots. These plots (also known as “bull’s eye” plots) are well established in nuclear cardiology and allow for direct segment- wise comparison of both modalities according to the standard 17-segment model of the left ventricle.
In the present study, we describe the procedure for the creation of such polar plots, validate the resulting plots against available standard methods in patients with myocardial infarction and provide examples that demonstrate the advantage of the novel approach over existing standards.
2. Methods
2.1. Patients
The University Hospital Essen ethics committee approved the examinations as part of fundamental research on integrated PET/MRI (registry number: 11-4825- BO). Inclusion criteria were: age of 18 years or older, no contraindications to the use of gadolinium-based contrast agents or general MR imaging, and a history of a successfully completed PET/MRI study. All patients gave written informed consent prior to the examination. PET/MRI was performed in 20 patients with acute myocardial infarction after interventional reperfusion (20 male, mean age: 51 [36 - 82] years, mean LVEF: 49 [27 - 66] %, time between event and PET/MRI scan: 10 [2 - 17] days). Three additional FDG PET/MRI examinations utilizing dietary suppression of myocardial glucose uptake in patients with myocardial inflammation (myocarditis: n = 2, cardiac sarcoidosis: n = 1) were evaluated to demonstrate the feasibility of the presented technique in cases of only focal myocardial tracer uptake.
2.2. PET/MRI Protocol
All scans were performed on an integrated whole-body 3T PET/MRI system (Biograph mMR, Siemens Healthcare, Erlangen, Germany) [5] . Examinations comprised the breath-hold acquisition of ECG-triggered segmented 2D inversion recovery Turbo-FLASH sequences (LGE) [6] with the following technical parameters: TI: 316 ± 47 [220 - 400] ms; TR: 5.66 ms; TE: 1.56 ms; flip-angle: 20˚; bandwidth: 465 Hz/Px; reconstructed pixel size: 1.3 × 1.3 mm2; slice thickness: 8 mm. Ten minutes before LGE imaging Gadobutrol (Gadovist®, Bayer Healthcare, Leverkusen, Germany) was administered as i.v. contrast agent at a weight-adapted dosage of 0.2 mmol/kg body weight. LGE imaging was performed in three long axes views (4 chamber view, 3 chamber view, 2 chamber view) and in contiguous short-axis views (8 mm slice thickness) covering the entire left ventricle.
In patients with myocardial infarction, resting myocardial glucose uptake was assessed by 2-deoxy-2-(18F)fluoro-D-glucose (FDG). Studies were performed after a fasting period of at least 6 h. One hour before tracer injection blood glucose level was checked and intravenous insulin was given in case of levels >150 mg/dl. After that 75 g glucose was orally administered. A mean activity of 172 ± 25 MBq FDG was administered intravenously. The PET/MRI scan started 102 ± 34 min after injection and comprised an electrocardiographically gated cardiac PET-scan with 3D image reconstruction (2 × 2 × 2 mm voxel size; 344 × 344 ma- trix). PET attenuation correction was performed using Dixon-based tissue segmentation [7] .
2.3. Image Processing
Volumetric PET data was resampled to match the slice thickness and orientation of the MRI acquisitions in short-axis views using OsiriX imaging software (OsiriX Foundation, Geneva, Switzerland). To reduce errors from miss-registration, corresponding LGE and PET slices were reregistered based on anatomical landmarks using OsiriX. The left ventricular myocardium (LVM) was segmented from LGE short-axis images using ImageJ (National Institute of Health, Bethesda, MD, USA) and exported as binary (1 and “not available”) pixel masks Ω:

with each short-axis image consisting of a matrix of pixels P(i,j) spanning i = 1 to L columns and j = 1 to M rows and with the upper left hand pixel as the coordinate system origin, i = j = 1.
LGE, PET and mask images were then imported into the R-software environment for statistical computing (R Foundation for Statistical Computing, Vienna, Austria) using the EBImage [8] and oro.dicom [9] packages. Image import and export operations were performed using lossless image encoding formats.
The area of the LVM was isolated on LGE and PET short-axis images by pixel-wise multiplication with the corresponding pixel masks:
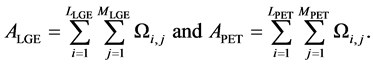
The center of the left ventricular blood pool (CLV) was defined as the center of gravity of the endocardial contour for each slice, defined with respect to the image origin.
Noise was reduced in the isolated LGE images ALGE using an edge-preserving 3 × 3 median filter. To reduce partial volume effects at the segmentation boundaries a second 3 × 3 median filter (including only on-mask pixels) was selectively applied to pixels being adjacent to a segmentation boundary. LVM areas showing late contrast-enhancement was delineated using an automated thresholding algorithm (ATA) based on an iterative selection method [10] . To avoid arbitrary thresholds in patients without LGE, regularization was added that required thresholds to be above the mean ALGE plus 2.5 standard deviations.
Based on the principle that myocardial cells undergo necrosis first in the subendocardium (wavefront phenomenon) [11] [12] , areas of microvascular obstruction (“no-reflow”) were defined as sub-threshold areas under LGE areas (in the direction of CLV) and automatically detected by an iterative radial search algorithm (Figure 1). The combination of thresholding and no-reflow detection yielded a classification mask Ω' for each slice:

Figure 1. Schematic outline of a transmural infarct in the anterolateral wall: (a) remote myocardium; (b) late gadolinium-enhancement; (c) no-reflow area. No-reflow areas are automatically detected based on the wave-front phenomenon. Therefore, hypointense areas located under hyperintense areas (in the epi-endocardial direction) are classified as no-reflow areas.
A radial sampling pattern of 90 wedge-shaped segments (every 4˚) perpendicular to the long-axis (with CLV being the center of rotation) was used in all slices equally spaced over the left ventricle, except for the most apical slice and the most basal slice (valvular plane) which were discarded because of partial volume effects. For each segment of ALGE the fraction of infarcted myocardium was calculated based on the mask Ω’. For each segment of APET the mean standardized uptake value normalized to patient body weight (SUVmean) was calculated and normalized to the maximum SUV of the entire left ventricle (SUVmax). The sampled data was then rendered into polar coordinate system plots where the most basal slice was projected onto the outmost orbital and the most apical slice onto the center of the plot, locating each pair of corresponding samples from ALGE and APET at identical coordinates. Before the rendering, color lookup tables were applied to the sampled data. Based on the continuous spectrum of SUVmean samples, a linear heat map ranging from “#0000FF” to “#FFFAFF” (in RGB encoding) was chosen for PET data. To account for the asymmetric bimodal distribution of the LGE fraction data with the majority of segments showing no late-enhancement (resulting in sample values of “0”), a biased color ramp palette ranging from “#4AA02C” to “#FF0000” was chosen for MRI data, yielding a wider color space on the right tail (the infarction areas) of the distribution.
2.4. Image Analysis
Manual segmentation of LGE areas was used as the standard of reference for the evaluation of the ATA classification. LGE areas, including hypointense no-ref- low zones, were manually delineated by two readers in consensus in each short-axis slice using ImageJ. During manual assessment both readers were blinded to the results of the ATA classification.
As the standard of reference for the evaluation of the MRI-segmentation based PET sampling pattern, standard polar plots were created with the Corridor4DM software package (INVIA Solutions, Ann Arbor, MI, USA). According to the 17-segment model of the American Heart Association (AHA), but excluding the apex (Segment 17), the relative tracer uptake was assessed for each myocardial segment in the corresponding polar plots and expressed in discrete levels from 1 to 5 (1: 1% - 20%, 2: 21% - 40%, 3: 41% - 60%, 4: 61% - 80%, 5: 81% - 100%; normalized to the overall maximum uptake of the left ventricular myocardium).
2.5. Statistical Analysis
Agreement between the manual LGE assessment and the ATA classification was analyzed using the Wilcoxon signed-rank test, Pearson product-moment correlation and Bland-Altman plots. Agreement between MRI-segmentation based PET polar plots and standard PET polar plots was assessed using intraclass correlation analysis and Cohen’s kappa with squared weighting.
3. Results
The software successfully produced co-registered LGE and PET polar plots from a testing set of 20 PET/MRI studies of myocardial infarction and a total of 196 short-axis slices (Figure 2). Depending on the size of the ventricle a total of 630 to 990 myocardial segments were evaluated in each patient (Figure 3).
Between manual and ATA segmentation no statistically significant difference in infarct sizes was found (p = 0.12). Mean (±standard deviation) infarct size according to manual segmentation was 17.5 ± 7.6 g (median = 15.3 g) and 17.0 ± 7.5 g (median = 14.6 g) according to ATA segmentation (Figure 4, left panel). Both methods were highly correlated (Pearson’s r = 0.96, p < 0.01; Figure 4, center panel). Bland-Altman analysis revealed lower and upper limits of agreement of −4.7 g and 3.6 g, respectively (Figure 4, right panel).
Agreement between MRI-segmentation based PET polar plots and standard PET polar plots was very high (Figure 5). Kappa analysis yielded a mean (± standard deviation) Cohen’s kappa of 0.91 ± 0.10 (p < 0.01 in all cases). Mean (± standard deviation) intraclass correlation coefficient was 0.95 ± 0.05 (p < 0.01 in
Figure 2. The upper row shows co-registered LGE (left) and PET (right) polar plots in a patient with a large transmural infarction due to occlusion of the left anterior descending coronary artery. The lower row shows co-registered LGE (left) and PET (right) polar plots in a patient with a large nearly transmural infarction due to occlusion of the left circumflex coronary artery.
Figure 3. This shows a PET/MRI examination in a 40-year-old patient with an acute occlusion of the circumflex artery and a consecutive transmural infarction of the inferolateral myocardium. The upper row displays 11 manually segmented short-axis slices acquired in late gadolinium-enhancement technique (right) and a basal short-axis slice without segmentation (left). The lower row displays 11 co-registered PET images (right) and a basal short-axis fusion of LGE and PET (left). The respective polar plots are displayed on the lower right of each row.
all cases).
In three additional patients with myocardial inflammation, the software successfully created polar plots that demonstrate the location and extent of myocardial inflammation with respect to the 17-segment model of the left ventricle (Figures 6-8).
4. Discussion
Twenty co-registered MRI-segmentation based LGE and PET polar plots were successfully created in patients with myocardial infarction. In LGE images, a high concordance regarding the visual depiction of the infarct area and the results of an automated threshold detection algorithm was found. Agreement between MRI-segmentation based PET polar plots and corresponding plots rendered with a standard software package for cardiac PET postprocessing was very high. This enables a visually and quantitatively consistent comparison of myocardial LGE and FDG uptake regarding the extent and location of myocardial infarction. Furthermore, the MR-segmentation based technique was demonstrated to create conclusive polar plots in PET/MRI studies of myocardial inflammation and dietary suppression of physiologic FDG uptake in the normal myocardium, that were not evaluable using standard software.
The good performance of the ATA in the detection of LGE matched the results of a recent study [13] that identified the Otsu algorithm [14] as the best method for fully automated thresholding. Ridler’s algorithm that was used in the present study, is very similar to Otsu’s algorithm. In fact, Ridler’s method can be described as an iterative version of Otsu’s method [15] . In addition to the previous study, our method could also detect and include no-reflow areas into the infarction zone, thus making it unnecessary to delineate these areas manually.
Figure 4. The left panel shows a boxplot comparing infarct sizes resulting from manual and automated late gadolinium enhancement detection (including no-reflow). No significant differences were found (Wilcoxon Test, p = 0.12). The scatterplot in the center panel indicates a high correlation between both methods (Pearson’s r = 0.96, p < 0.01). The Bland-Altman plot in the right panel reveals limits of agreement between both methods from −5.3 g to 3.7 g.
Despite being based on a different sampling pattern, the MRI-segmentation based PET polar plots were very similar to corresponding standard plots, where the segmentation was based on the detection of the mid-myocardial iso-surface [16] .
We chose the polar map as a common data representation because it allows for rapid quantification of myocardial pathologies with respect to the 17-seg- ment model of the left ventricle, gives a fast and intuitive overview and is commonly used in nuclear cardiology. However, currently available software for the creation of left ventricular polar plots was originally designed for non-hybrid imaging applications in nuclear cardiology. These methods perform the segmentation of the left ventricular myocardial boundaries on functional (PET or
Figure 5. Both panels display polar plots of the same cardiac PET dataset in a patient with an acute occlusion of the circumflex artery and consecutive myocardial infarction. The left panel displays a MRI-segmentation based PET polar plot rendered with our software. The right panel displays a PET polar plot rendered with a standard software package for cardiac PET postprocessing (Corridor4DM, INVIA Solutions, Ann Arbor, MI, USA). Apart from different color encodings, both plots demonstrate high agreement between both approaches.
Figure 6. FDG PET/MR images in a patient with myocarditis and atypical, infarct-like, nearly transmural late gadolinium-enhancement in the basal septum (top left image). The patient was prepared with a high-fat low-carbohydrate protein-permitted diet shifting metabolism in the normal myocardium to the utilization of free fatty acids. As intended, significant FDG uptake is only observed in segments infiltrated by inflammatory cells. The left ventricular myocardium has no sufficient visual representation in PET images (bottom left: axial PET image; bottom right: coronal PET image). Consequently, PET polar plots cannot be created by conventional approaches that rely on the segmentation of the myocardium in PET image space. Co-registered polar plots of late gadolinium-en- hancement (top center) and PET (top right) were created using the MRI-segmentation based approach described in the present paper. Both plots demonstrate high agreement between late gadolinium-enhancement (according to myocardial necrosis) and FDG uptake (according to inflammatory activity).
SPECT) imaging data and thus strongly depend on relatively homogeneous and distinctive myocardial uptake of the respective tracer. In cases of specific tracers against certain imaging targets (like inflammatory cells) or poor myocardium to
Figure 7. FDG PET/MR images in a patient with cardiac sarcoidosis. PET images (top left, bottom left) as well as fused PET/MRI (top right) demonstrate focal FDG uptake in the inferolateral left ventricular wall. The extent and location of myocardial inflammation is visualized in one comprehensive polar plot (bottom right) created by the technique presented in this paper.
background ratio, polar plots cannot be created using these methods. In this respect, it is important to appreciate that some of the most promising applications of cardiac PET/MRI involve the imaging of myocardial inflammation, which currently is either performed using FDG with dietary suppression of myocardial glucose uptake [17] or with specific tracers against inflammatory cells that do not show significant uptake in myocardial cells [18] . In both cases it is not possible to create valid polar plots of the left ventricular tracer uptake using standard software. This could also happen in extensive transmural infarction where large parts of the left ventricle may be invisible in metabolic PET images. On the other hand, the standard approach of polar map generation from PET data has the advantage of being not dependent on co-registered morphological (MRI or CT) images and a high degree of automation if the respective tracer uptake demonstrates a strong myocardium to background ratio [19] .
While polar plots are commonly used in nuclear cardiology, these plots are only sporadically used in cardiac MRI today [20] [21] . Because co-registered morphological MR images are by default available in PET/MRI, we performed segmentation of myocardial boundaries in MR images. This provides not only a straightforward workaround in cases of sparse myocardial tracer uptake but also results in inherently co-registered polar plots if MR imaging findings like late gadolinium-enhancement are mapped to a polar plots as well. Utilizing the same
Figure 8. FDG PET/MR images in another patient with myocarditis. PET images (top left, bottom left) as well as fused PET/MRI (top right) demonstrate inhomogeneous FDG uptake in the septum and the anterior wall. The extent and location of myocardial inflammation is visualized in one comprehensive polar plot (bottom right) created by the technique presented in this paper.
underlying MRI-based segmentation and a common sampling scheme improves the spatial coherence of the resulting polar plots, which supports the ultimate goal of an integrated assessment of PET/MRI studies. Furthermore, this technique should allow for a more precise segmentation of myocardial boundaries as MR images provide high anatomical resolution and excellent soft tissue contrast. This novel approach was evaluated in a patient cohort where the classic PET- only approach was applicable as well, which allowed for the comparison between both. However, after the feasibility of MRI-based PET segmentation has been shown, this can be applied to all kinds of cardiac PET/MRI examinations, as it is completely independent of the available PET data as we have demonstrated in FDG PET/MRI studies of myocardial inflammation, where dietary suppression of myocardial glucose uptake was performed. In future, this might improve the quantification of findings in hybrid cardiac imaging studies, which will become even more important with the clinical availability of highly specific (pathology targeted) PET tracers.
Since the attachment of the right ventricular wall to the left ventricle was not incorporated into our sampling model, a rotational deviation with regard to the 17-segment model may occur. Alignment to the 17-segment model of the AHA was performed by manual rotation in the present study, however, automated alignment could be easily integrated into a future version of the software. Although both modalities were simultaneously acquired using ECG triggering/ gating and MR acquisitions were performed in breath-hold, some misalignments still had to be corrected with anatomical landmarks-based registration. Therefore, minor errors on the quantitative level resulting from imperfect co-registra- tion have to be assumed. Segmentation of myocardial boundaries was performed on the pixel level. In future, accuracy could potentially be increased by using subpixel segmentation. While manual segmentation of myocardial boundaries and automated LGE (including no-reflow) detection yielded excellent results, this must at least partially be attributed to the overall high quality of LGE images on this PET/MRI device that was described before [1] . The reproducibility under less optimal conditions still has to be demonstrated. Finally, the most apical and basal short-axis slices were discarded due to partial volume effects in 2-dimen- sional MR images. Further development of that method could at least partially compensate for this by integrating long-axis LGE images, reducing the slice thickness of the LGE short-axis images or ultimately using a 3-dimensional sampling pattern, which however would also require 3-dimensional LGE data.
5. Conclusion
In conclusion, we have described a straightforward and robust software approach for the creation of co-registered PET and MRI polar plots of cardiac PET/MRI studies that work independently of myocardial tracer uptake and thus are even feasible in studies with sparse myocardial tracer uptake like PET/MRI imaging of myocardial inflammation.
Cite this paper
Nensa, F., Poeppel, T.D., Tezgah, E., Heusch, P., Nassenstein, K., Forsting, M., Bockisch, A., Erbel, R. and Schlosser, T. (2017) Integrated Assessment of Cardiac PET/MRI: Co-Registered PET and MRI Polar Plots by Mutual MR-Based Seg- mentation of the Left Ventricular Myocardium. World Journal of Cardiovascular Di- seases, 7, 91-104. https://doi.org/10.4236/wjcd.2017.74010
References
- 1. Nensa, F., Poeppel, T.D., Beiderwellen, K., Schelhorn, J., Mahabadi, A.A., Erbel, R., et al. (2013) Hybrid PET/MR Imaging of the Heart: Feasibility and Initial Results. Radiology, 268, 366-373.
https://doi.org/10.1148/radiol.13130231 - 2. White, J.A., Rajchl, M., Butler, J., Thompson, R.T., Prato, F.S. and Wisenberg, G. (2013) Active Cardiac Sarcoidosis: First Clinical Experience of Simultaneous Positron Emission Tomography-Magnetic Resonance Imaging for the Diagnosis of Cardiac Disease. Circulation, 127, e639-e641.
https://doi.org/10.1161/CIRCULATIONAHA.112.001217 - 3. Ibrahim, T., Nekolla, S.G., Langwieser, N., Rischpler, C., Groha, P., Laugwitz, K.-L., et al. (2012) Simultaneous Positron Emission Tomography/Magnetic Resonance Imaging Identifies Sustained Regional Abnormalities in Cardiac Metabolism and Function in Stress-Induced Transient Midventricular Ballooning Syndrome. Circulation, 126, e324-e326.
https://doi.org/10.1161/CIRCULATIONAHA.112.134346 - 4. Schlosser, T., Nensa, F., Mahabadi, A.A. and Poeppel, T.D. (2013) Hybrid MRI/PET of the Heart: A New Complementary Imaging Technique for Simultaneous Acquisition of MRI and PET Data. Heart, 99, 351-352.
https://doi.org/10.1136/heartjnl-2012-302740 - 5. Delso, G., Fürst, S., Jakoby, B., Ladebeck, R., Ganter, C., Nekolla, S.G., et al. (2011) Performance Measurements of the Siemens mMR Integrated Whole-Body PET/MR Scanner. Journal of Nuclear Medicine, 52, 1914-1922.
https://doi.org/10.2967/jnumed.111.092726 - 6. Lin, D. and Kramer, C.M. (2008) Late Gadolinium-Enhanced Cardiac Magnetic Resonance. Current Cardiology Reports, 10, 72-78.
https://doi.org/10.1007/s11886-008-0014-4 - 7. Martinez-Möller, A., Souvatzoglou, M., Delso, G., Bundschuh, R.A., Chefd’hotel, C., Ziegler, S.I., et al. (2009) Tissue Classification as a Potential Approach for Attenuation Correction in Whole-Body PET/MRI: Evaluation with PET/CT Data. Journal of Nuclear Medicine, 50, 520-526.
https://doi.org/10.2967/jnumed.108.054726 - 8. Pau, G., Fuchs, F., Sklyar, O., Boutros, M. and Huber, W. (2010) EBImage—An R Package for Image Processing with Applications to Cellular Phenotypes. Bioinformatics, 26, 979-981.
https://doi.org/10.1093/bioinformatics/btq046 - 9. Whitcher, B., Schmid, V.J. and Thorton, A. (2011) Working with the DICOM and NIfTI Data Standards in R. Journal of Statistical Software, 44, 1-29.
https://doi.org/10.18637/jss.v044.i06 - 10. Ridler, T.W. and Calvard, S. (1978) Picture Thresholding Using an Iterative Selection Method. IEEE Transactions on Systems, Man, and Cybernetics, 8, 630-632.
https://doi.org/10.1109/TSMC.1978.4310039 - 11. Reimer, K.A., Lowe, J.E., Rasmussen, M.M. and Jennings, R.B. (1977) The Wave-Front Phenomenon of Ischemic Cell Death. 1. Myocardial Infarct Size vs. Duration of Coronary Occlusion in Dogs. Circulation, 56, 786-794.
https://doi.org/10.1161/01.CIR.56.5.786 - 12. Skyschally, A., Schulz, R. and Heusch, G. (2013) Pathophysiology of Myocardial Infarction: Protection by Ischemic Pre- and Post-conditioning. Herz Kardiovaskuläre Erkrankungen, 33, 88-100.
https://doi.org/10.1007/s00059-008-3101-9 - 13. Vermes, E., Childs, H., Carbone, I., Barckow, P. and Friedrich, M.G. (2013) Auto-Threshold Quantification of Late Gadolinium Enhancement in Patients with Acute Heart Disease. Journal of Magnetic Resonance Imaging, 37, 382-390.
https://doi.org/10.1002/jmri.23814 - 14. Otsu, N. (1979) A Threshold Selection Method from Gray-Level Histograms. IEEE Transactions on Systems, Man, and Cybernetics, 9, 62-66.
https://doi.org/10.1109/TSMC.1979.4310076 - 15. Xue, J.-H. and Zhang, Y.-J. (2012) Ridler and Calvard’s, Kittler and Illingworth’s and Otsu’s Methods for Image Thresholding. Pattern Recognition Letters, 33, 793-797.
- 16. Nekolla, S.G., Miethaner, C., Nguyen, N., Ziegler, S.I. and Schwaiger, M. (1998) Reproducibility of Polar Map Generation and Assessment of Defect Severity and Extent Assessment in Myocardial Perfusion Imaging Using Positron Emission Tomography. European Journal of Nuclear Medicine, 25, 1313-1321.
https://doi.org/10.1007/s002590050301 - 17. Nensa, F., Tezgah, E., Schweins, K., Goebel, J., Heusch, P., Nassenstein, K., et al. (2016) Evaluation of a Low-Carbohydrate Diet-Based Preparation Protocol without Fasting for Cardiac PET/MR Imaging. Journal of Nuclear Cardiology, 1-9.
https://doi.org/10.1007/s12350-016-0443-1 - 18. Rischpler, C., Nekolla, S.G., Kossmann, H., Dirschinger, R.J., Schottelius, M., Hyafil, F., et al. (2015) Upregulated Myocardial CXCR4-Expression after Myocardial Infarction Assessed by Simultaneous GA-68 Pentixafor PET/MRI. Journal of Nuclear Cardiology, 23, 131-133.
https://doi.org/10.1007/s12350-015-0347-5 - 19. Laubenbacher, C., Rothley, J., Sitomer, J., Beanlands, R., Sawada, S., Sutor, R., et al. (1993) An Automated Analysis Program for the Evaluation of Cardiac PET Studies: Initial Results in the Detection and Localization of Coronary Artery Disease Using Nitrogen-13-Ammonia. Journal of Nuclear Medicine, 34, 968-978.
- 20. Termeer, M., Oliván Bescós, J., Breeuwer, M., Vilanova, A., Gerritsen, F. and Gröller, E. (2007) CoViCAD: Comprehensive Visualization of Coronary Artery Disease. IEEE Transactions on Visualization and Computer Graphics, 13, 1632-1639.
https://doi.org/10.1109/TVCG.2007.70550 - 21. Noble, N.M., Hill, D.L., Breeuwer, M. and Razavi, R. (2004) The Automatic Identification of Hibernating Myocardium. In: Barillot, C., Haynor, D.R. and Hellier, P., Eds., Medical Image Computing and Computer-Assisted Intervention, Lecture Notes in Computer Science, Vol. 3217, Springer, Berlin, Heidelberg, 890-898.
https://doi.org/10.1007/978-3-540-30136-3_108









