Open Journal of Molecular and Integrative Physiology
Vol.3 No.2(2013), Article ID:31298,5 pages DOI:10.4236/ojmip.2013.32008
Protein phosphatase 4 is involved in the late development of Dictyostelium discoideum*
![]()
Department of Agricultural Science, Kinki University, Nara, Japan
Email: #yoshida_m@nara.kindai.ac.jp
Copyright © 2013 Ichiro Kamei et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received 29 January 2013; revised 1 March 2013; accepted 10 March 2013
Keywords: Dictyostelium; Protein Phosphatase; Fruiting-Body Formation; Knockout Mutant
ABSTRACT
A cDNA clone SSJ337 (accession no. AF161253) of 1230 bp, encoding a catalytic subunit of protein phosphatase 4, was selected as one of the clones expressed specifically in prestalk cells from a cDNA library of D. discoideum slugs. Cells transformed with a knockout construct of SSJ337 showed an aberrant and tiny fruiting-body formation with a short stalk. A knockout mutant, SSJ337KO was allowed to develop much slower than a wild-type AX2 after the post-aggregation stage. This suggested that the SSJ337 cDNA clone has played an important role especially in the later development of Dictyostelium discoideum. Results from Northern blotting, analysis showed that transcripts for SSJ337 were accumulated at 16 h to 24 h after starvation began.
1. INTRODUCTION
Dictyostelium discoideum is a unique organism having a single amoeba stage and a multicellular stage in its life cycle. D. discoideum has been used as a model organism to study development or differentiation mechanisms. After starvation, D. discoideum cells aggregate at 8 h and eventually form a fruiting body consisting of spore and stalk cells at 24 h [1].
The knockout mutants for genes involved in the development of D. discoideum show aberrant development. A cAMP receptor subtype 2, CAR2, is required for the cAMP-directed sorting of prestalk cells during pattern formation of an aggregation mound, and the knockout mutant cells remain in the tight aggregation stage [2]. A MAP kinase, ERK2, is involved in cAMP-dependent signal transduction during aggregation and the cell-type differentiation process, and the knockout mutant cells do not aggregate and lose the ability to activate adenylyl cyclase in response to extracellular cAMP [3]. A signal transducer and an activator of transcription, Dd-STATa, is a repressor for stalk-cell differentiation, and the knockout mutant cells remain at the slug stage before forming small spore masses [4]. A Dictyostelium cDNA project [5] and a genome project [6] have been performed and a transcriptional profile of multicellular formation has been analyzed by DNA microarray [7]; however, the functions of many individual genes remain unclear. Therefore, we carried out gene disruption analysis using cDNA clones expressed specifically in prestalk cells from the Dictyostelium slug cDNA library to identify the genes involved in the fruiting-body formation of D. discoideum [8]. Among these cDNA clones, we studied the function of a SSJ337 cDNA clone, encoding a catalytic subunit of protein phosphatase 4.
2. MATERIALS AND METHODS
2.1. Cell Culture and Development
Wild-type AX2 was cultivated at 22˚C in HL5 medium. A knockout mutant for a SSJ337 cDNA clone, SSJ337KO, was grown in HL5 medium containing 10 μg/ml Blasticidin S (Funakoshi, Tokyo, Japan). Development was begun by washing the cells in 17 mM phosphate buffer, pH 6.1. The cells were suspended in the same buffer at a density of 1 × 107/ml. They were agitated on a rotary shaker at 150 rpm, or spread on agar plates, and incubated at 22˚C for the indicated times as reported previously [9].
2.2. Preparation of a Knockout Construct
A knockout construct for the SSJ337 cDNA clone (accession no. AF161253), which was primarily sequenced by us as a clone of the “Dictyostelium cDNA project in Japan, http://www.csm.biol.tsukuba.ac.jp/cDNAproject.html”, was prepared as follows (Figure 1). A BSR cassette containing bsr (blasticidin-resistant gene) [10] was inserted into the Xba I site of SSJ337cDNA in pBluescript KS (-), followed by PCR-mediated amplification using rTaq DNA polymerase, T3 and T7 primers (Merck, Darmstadt, Germany). PCR products were purified using QIAGENtip 20 (QIAGEN, Duesseldorf, Germany). The knockout construct was transformed into AX2 cells by electroporation, and transformants were isolated as described previously [8,11]. In brief, AX2 cells were suspended in electroporation buffer (10 mM Na2HPO4/KH2PO4, 50 mM sucrose, pH 6.1) to a density of 1.5 × 107 cells/ml and 10 μg of DNAs was added to the cell suspension. D. discoideum AX2 cells were transformed with a Bio-Rad gene pulser 1 - 1.5 kV/0.4 cm, 3 μF. After 24 h, 10 μg/ml Blasticidin S was added. After 10 - 12 days in the presence of the antibiotic, three transformants were independently isolated.
2.3. Northern Blotting Analysis and Southern Blotting Analysis
A cDNA probe was labeled with digoxigenin (DIG) by the random hexamer procedure using a DIG DNA labeling kit (Roche Diagnostics, Mannheim, Germany). Genomic DNAs and total RNAs were extracted with ISOGEN (Nippon gene, Japan). Total RNAs were extracted from wild-type AX2 cells starved for 0, 8, 16 or 24 h on non-nutrient agar plates, and Northern blotting analysis was conducted using SSJ337 cDNA as a probe. Ten μg of total RNAs was separated on formaldehydeagarose gel. Five μg of genomic DNAs was digested with an appropriate restriction enzyme and separated on agarose gel. After agarose gel electrophoresis, agarose gels were transferred onto HybondTMN+nylon membrane (GM Healthcare, Buckinghamshire, UK) with VacuGeneXL (GM Healthcare). Hybridization was carried out at 50˚C in DIG easy hyb (Roche Diagnostics) for Southern blotting analysis and at 45˚C for Northern blotting analysis.
3. RESULTS AND DISCUSSION
3.1. Isolation of a Knockout Mutant for SSJ337 cDNA Clone
To identify novel genes involved in fruiting-body forma-
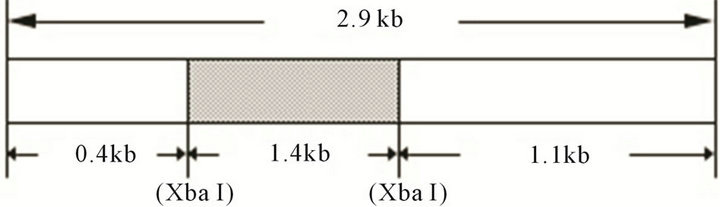
Figure 1. A knockout construct, SSJ337-Bsr, showing that BSR cassette is inserted into Xba I site of SSJ337 cDNA.
tion, we prepared knockout constructs by inserting a BSR cassette into each appropriate restriction-enzyme site of several dozens cDNA expressed specifically in prestalk cells from a Dictyostelium discoideum slug cDNA library. Of them, a knockout construct for SSJ337 cDNA clone (SSJ337-Bsr) was prepared by inserting the BSR cassette into Xba I of SSJ337 cDNA. Three knockout mutants transformed with SSJ337-Bsr (SSJ337KO) and isolated independently, showed the same aberrant and tiny fruiting bodies. In particular, the stalks of transformants were much shorter than those of wild-type AX2. Southern blotting analysis of transformants, using SSJ337 cDNA labeled with DIG as a probe, indicated that a single band of 6.3 kb in the wild-type AX2 shifted to 7.7 kb in transformants. In addition, a single band of 3.1 kb in the wild-type AX2 shifted to 4.5 kb in transformants (Figure 2(a)). After stripping a probe off a membrane, the membrane was incubated with bsr (blasticidin-resistant gene) labeled with DIG. As a result, no signal was observed in wild-type AX2, but a single band of 7.7 kb or 4.5 kb was observed in transformants (Figure 2(b)). Northern blotting analysis using total RNAs from cells starved for 16 h on non-nutrient agar plates showed no detectable signals in transformants (Figure 2(c)). Results from Southern blotting and Northern blotting analysis suggested that the knockout construct was inserted into chromosomal DNAs of transformants by homologous recombination, and that the insertion led to the disruption of a single copy gene of genomic DNAs corresponding to the SSJ337 cDNA clone.
3.2. Functional Analysis of SSJ337 cDNA Encoding a Putative Protein Phosphatase 4 Catalytic Subunit
To analyze the function of the SSJ337 cDNA clone, SSJ337KO cells were allowed to develop on agar plates, and their morphology was observed. Until the formation of a tight aggregation during development, there were no differences between wild-type AX2 and SSJ337KO cells (Figures 3(a) and (b)). After the post-aggregation stage, the development of SSJ337KO cells became much slower than that of wild-type AX2 cells. Migrating slugs of wild-type AX2 cells appeared at 16 h and mature fruiting bodies formed at 24 h (Figures 3(c) and (e)), while SSJ337KO cells reached the finger stage at 16 h and formed aberrant and tiny fruiting bodies at 36 h after starvation began (Figures 3(d) and (f)).
Transcript levels of the SSJ337 cDNA clone during development were investigated by Northern blotting analysis. Signals were detected in cells starved for 0 to 24 h (Figure 4). Among them, strong signals were observed during late development from 16 to 24 h. Taken together with the phenotype of SSJ337KO, these results suggested that the protein encoded by SSJ337 cDNA
 (a)
(a) (b)
(b) (c)
(c)
Figure 2. Southern blotting and Northern blotting analysis of a knockout mutant for SSJ337 cDNA (SSJ337KO). Southern blotting analysis for wild-type AX2 (Co) and SSJ337KO cells (T1, T2) was conducted. Genomic DNAs extracted from AX2 and SSJ337KO cells were digested with EcoR I or EcoR V. They were blotted onto a membrane that was incubated with SSJ337 cDNA (a) or bsr fragment (b) labeled with DIG. M, λ DNA digested with Hind III. In (c), Northern blot analysis for cells starved for 16 h was conducted. Ten mg of total RNAs was separated on formaldehyde-agarose gel and blotted onto a membrane that was incubated with SSJ337 cDNA labeled with DIG (left panel). An agarose gel was stained with with EtBr to confirm that the same amount of total RNA was applied to each lane (right panel).
 (a)
(a) (b)
(b) (c)
(c)
Figure 3. Morphology of wild-type AX2 and SSJ337KO cells. Wild-type AX2 and SSJ337KO cells were starved for the indicated times on non-nutrient agar plates. (a) AX2 cells starved for 8 h; (b) SSJ337KO cells starved for 8 h; (c) AX2 cells starved for 16 h; (d) SSJ337KO cells starved for 16 h; (e) AX2 cells starved for 24 h and mature fruiting bodies formed at 24 h; (f) SSJ337KO cells starved for 36 h and tiny fruiting bodies formed at 36 h. The scale bar indicates 50 μm.
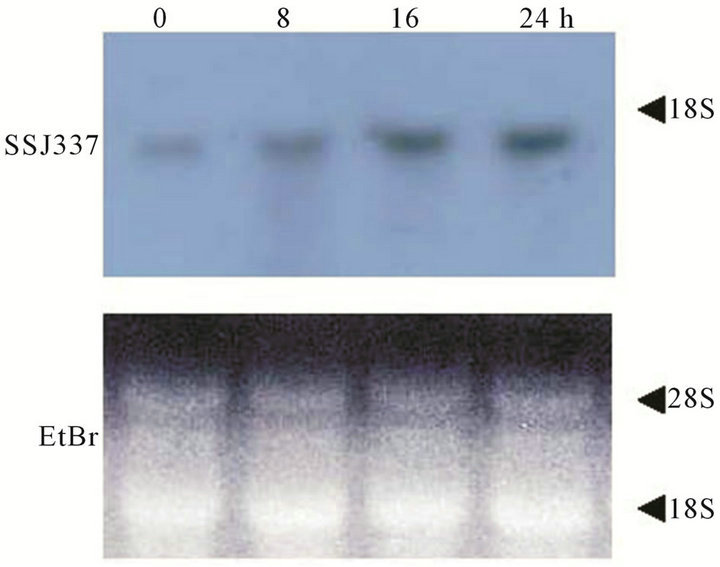
Figure 4. Transcripts for SSJ337 in wild-type AX2 cells. Ten mg of total RNAs was isolated from cells starved for the indicated times (0 - 24 h). Total RNAs were separated on formaldehyde-agarose gel and blotted onto a membrane that was incubated with SSJ337 cDNA labeled with DIG (upper panel). An agarose gel was stained with with EtBr to confirm that the same amount of total RNA was applied to each lane (lower panel).
played an important role in the morphogenesis of late development.
Partial DNA nucleotide sequences of the SSJ337 cDNA clone have been determined by the Dictyostelium cDNA project in Japan. A homology search was conducted to identify the protein encoded by the SSJ337 cDNA clone. The nucleotide sequences of the SSJ337 cDNA clone completely corresponded to those of the protein phosphatase 4 catalytic subunit (PP4C, accession no. AF161253). The phosphorylation of serine/threonine residues on proteins by protein kinases is responsible for the communication of many intracellular signals [12]. Moreover, protein dephosphorylation by serine/threonine phosphatase is important in the on/off switch of phosphorylation [12]. According to substrate specificity, inhibitor sensitivity, and requirement for divalent cations, serine/threonine phosphatases have been classified in three families: PPP family, PPM family and FCP family. PPP family consists of four subfamilies: PPP1 subfamily, PPP2/4/6 subfamily, PPP3 subfamily and PPP5/7 subfamily [13-15]. In D. discoideum, there is an abundance of biochemical and molecular biological information about protein kinases, but little is known about serine/ threonine phosphatases. D. discoideum PP1 and PP2A phosphatases have been identified [16], and D. discoideum PP2A 65 kDa A-subunit and 55 kDa B-subunit have been isolated [17]. Partial cDNA sequences of several predictable phosphatases have been identified as expressing sequence tags through the Dictyostelium cDNA project in Japan; however, the cellular functions of these many phosphatases remain unknown. PP4 is a member of PPP2/4/6 subfamily. The catalytic subunit of PP4 (PP4C) has 65% identity with the catalytic subunit of PP2A in amino acid sequences [18]. PP4 and PP2A regulate Hedgehog signal transduction of Drosophila [19]. PP4 is highly conserved among many mammals in addition to having 99% identity between human and rabbit in amino acid sequences [20]. PP4C is predominantly localized in the nucleus of rat brain and testis, and it is most highly expressed in the testis [21]. PP4 has been demonstrated to be an essential enzyme in the development of Drosophila embryos [22]. The results of the SSJ337KO phenotype and Northern blotting analysis of SSJ337 cDNA suggested that the protein encoded by this gene played an important role in the morphogenesis of late development of D. discoideum.
4. ACKNOWLEDGMENTS
One of authors (M. Y.) was a member of the “Dictyostelium cDNA project team in Japan”. This study was supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan, No. 18580341, and by “Academic Frontier” Project for Private Universities: matching fund subsidy from Ministry of Education, Culture, Sports, Science and Technology.
REFERENCES
- Bonner, J.T. (1982) Comparative biology of cellular slime molds. In: Loomis, W.F., Ed., The Development of Dictyostelium discoideum, Academic Press, New York, 1-28.
- Saxe, C.L.III, Ginsburg, G.T., Louis, J.M., Johnson, R.L., Devreotes, P.N. and Kimmel, A.R. (1993) CAR2, a prestalk cAMP receptor required for normal tip formation and late development of Dictyostelium discoideum. Genes & Development, 7, 262-272. doi:10.1101/gad.7.2.262
- Segall, J.E., Kuspa, A., Shaulsky, G., Ecke, M., Maeda, M., Gaskins, C., Firtel, R.A. and Loomis, W.F. (1995) A MAP kinase necessary for receptor-mediated activation of adenylyl cyclase in Dictyostelium. The Journal of Cell Biology, 128, 405-413. doi:10.1083/jcb.128.3.405
- Mohanty, S., Jermyn, K.A., Early, A., Kawata, T., Aubry, L., Ceccarelli, A., Schaap, P., Williams, J.G. and Firtel, R.A. (1999) Evidence that the Dictyostelium Dd-STATa protein is a repressor that regulates commitment to stalk cell differentiation and is also required for efficient chemotaxis. Development, 126, 3391-3405.
- Morio, T., Urushihara, H., Saito, T., Ugawa, Y., Mizuno, H., Yoshida, M., Yoshino, R., Mitra, B.N., Pi, M., Sato, T., Takemoto, K., Yasukawa, H., Williams, J., Maeda, M., Takeuchi, I., Ochiai, H. and Tanaka, Y. (1998) The Dictyostelium development cDNA project: Generation and analysis of expressed sequence tags from the first-finger stages of development. DNA Research, 5, 335-340. doi:10.1093/dnares/5.6.335
- Eichinger, L., Pachebat, J.A., Gloeckner, G., Rajandream, M.-A., Sucgang, R., Berriman, M., Song, J., Olsen, R., Szafranski, K., Xu, Q., et al. (2005) The genome of the social amoeba Dictyostelium discoideum. Nature, 435, 43- 57. doi:10.1038/nature03481
- Van Driessche, N., Shaw, C., Katoh, M., Morio, T., Sucgang, R., Ibarra, M., Kuwayama, H., Saito, T., Urushihara, H., Maeda, M., Takeuchi, I., Ochiai, H., Eaton, W., Tollett, J., Halter, J., Kuspa, A., Tanaka, Y. and Shaulsky, G. (2002) A transcriptional profile of multicellular development in Dictyostelium discoideum. Development, 129, 1543-1552.
- Takamoto, K., Ootaki, T. and Yoshida, M. (2001) Identification of genes involved in fruiting-body formation of Dictyostelium discoideum. Bulletin of the Institute for Comprehensive Agricultural Sciences Kinki University, 9, 123-127.
- Yoshida, M., Ishida, S. and Iizuka, Y. (1991) Mutants of Dictyostelium discoideum with altered carbohydrate moieties of contact site A. Cell Structure Function, 16, 383- 390. doi:10.1247/csf.16.383
- Sutoh, K. (1993) A transformation vector for Dictyostelium discoideum with a new selectable marker bsr. Plasmid, 30, 150-154. doi:10.1006/plas.1993.1042
- Sakuragi, N., Ogasawara, N., Tanesaka, E. and Yoshida, M. (2005) Functional analysis of a novel gene, DD3-3, from Dictyostelium discoideum. Biochemical Biophysical Research Communications, 331, 1201-1206. doi:10.1016/j.bbrc.2005.03.244
- Virshup, D. and Shenolikar, M. (2009) From promiscuity to precision: Protein phosphatases get a makeover. Molecular Cell, 33, 537-545. doi:10.1016/j.molcel.2009.02.015
- Cohen, P.T.W. (2004) Overview of protein serine/threonine phosphatases. In: Arino, J. and Alexander, D.R., Eds., Protein phosphatases. Topics in Current Genetics, 5, Springer-Verlag, Berlin, 1-15. doi:10.1007/978-3-540-40035-6_1
- Depaoli-Roach, A.A., Park, I.K., Cerovsky, V., Csotos, C., Durbin, S.D., Kuntz, M.J., Sitikov, A., Tang, P.M., Verin, A. and Zolnierowicz, S. (1994) Serine/threonine protein phosphatases in the control of cell function. Advances in Enzyme Regulation, 34, 199-224. doi:10.1016/0065-2571(94)90017-5
- Mumby, M.C. and Walter, G. (1993) Protein serine/threonine phosphatases: Structure, regulation, and functions in cell growth. Physiological Reviews, 73, 673-699.
- Haribabu, B. and Dottin, R.P. (1991) Homology cloning of protein kinase and phosphoprotein phosphatase sequences of Dictyostelium discoideum. Developmental Genetics, 12, 45-49. doi:10.1002/dvg.1020120109
- Murphy, M.B., Levi, S.K. and Egelhoff, T.T. (1999) Molecular characterization and immunolocalization of Dictyostelium discoideum protein phosphatase 2A. FEBS Letters, 456, 7-12. doi:10.1016/S0014-5793(99)00835-2
- Brewis, N.D., Street, A.J., Prescott, A.R. and Cohen, P.T. W. (1993) PPX, a novel protein serine/threonine phosphatase localized to centrosomes. EMBO Journal, 12, 987- 996.
- Jia, H., Liu, Y., Yan, W. and Jia, J. (2009) PP4 and PP2A regulate Hedgehog signaling by controlling Smo and Ci phosphorylation. Development, 136, 307-316. doi:10.1242/dev.030015
- Brewis, N.D. and Cohen, P.T.W. (1992) Protein phosphatase X has been highly conserved during mammalian evolution. Biochimica Biophysica Acta (BBA)—Gene Structure and Expression, 1171, 231-233. doi:10.1016/0167-4781(92)90129-N
- Kloeker, S., Bryant, J.C., Strack, S., Colbran, R.J. and Wadzinski, B.E. (1997) Carboxymethylation of nuclear protein serine/threonine phosphatase X. Biochemical Journal, 327, 481-486.
- Helps, N.R., Brewis, N.D., Lineruth, K., David, T., Kaiser, K. and Cohen, P.T. (1998) Protein phosphatase 4 is an essential enzyme required for organisation of microtubules at centrosomes in Drosophila embryos. Journal of Cell Science, 111, 1331-1340.
NOTES
*This paper has been submitted to FEMS Lett. (2006) before publication of Mol. Cell Biol. 27: 3817-3827 (2007).
#Corresponding author.

