DNA Sequencing Modified Method through Effective Regulation of Its Translocation Speed in Aqueous Solution ()
1. Introduction
The process of DNA sequencing is a precise determination of the amount and distribution of the nucleotides in DNA molecules. Quick improvements in the cost and speed of DNA sequencing are having a strong influence on comprehensive genome research. Methods of DNA sequencing and their application are detailed analyzed in [1]. In the past decade, the single-nanometer-scale pores demonstrated great capability for the detection, identification, and characterization of DNA [2] [3] and RNA [4] [5]. In recent years, rapid advances have been made and many construction’s architectures have been put forward for novel approaches to bio-molecular sensing using nanoelectronics, including the advent of tunnel junctions as a sensing platform. Within the past decade, nanogap electrodes have attracted a lot of attention because of their potential applications in the progressive miniaturization of electronics and as nanoscale tool for analysis of single molecular properties. DNA bases can be identified statistically in nanopore translocation events. Nanogap electrodes can be simply defined as a pair of electrodes with a gap that can be measured in nanometers. The transport dynamics of the charged molecules in the nanosize constructions located in aqueous solution environment is the result of multiple factors including, electrostatic and hydrodynamic interactions, drift and diffusion.
One major challenge of nanopore-based DNA sequencing technology is to find an efficient way to reduce DNA translocation speed. It is necessary that each nucleotide can reside long enough in the measuring pore for creation molecular junctions. Magnitudes of those current will help identification of nucleotides [6]. In order to take control of the DNA molecule translation process, various theories and approaches have been put forward in recent decades.
Authors of Ref. [7] considered that the reasonable value of the DNA translocation speed is near to 0.01 - 1 ms per base, which is equivalent to (10 - 1000) bases/s. Peng and Ling reversed the DNA translocation and achieved an average speed of 0.0096 bases/μs = 9600 bases/s [8]. As the translocation times depend strongly on the nucleotide type [9], imply that polymer-pore interactions, rather than the more generic hydrodynamic drag, play an important role in determining the translocation dynamics. The interaction of the polymer with pores is described in detail in [9]. In [10], the authors propose a feedback device architecture for regulating DNA translocation by modulating the effective surface charge density of a nanopore wall. It was shown that the rate of DNA movement can be reduced at a rate of about 55 mm/s per 1 mV/nm. The review [11] focuses on a single aspect in the transport dynamics of a polymer drawn inside a nanoscopic channel. Primarily, the dynamics of polynucleotides is discussed. Some of the concepts that are discussed in [11] apply to uncharged polymers.
Thus, a key challenge to DNA sequencing with nanopores is to find methods to slow down and control DNA translocation. DNA translocation speeds can be reduced somewhat by decreasing temperature [12] [13], or increasing solvent viscosity [7] [14], but these methods do not reduce the variations in the translocation dynamics because of DNA-pore interactions [15] [16] [17] [18]. Problems of controlling DNA motion and translocation in a nanopore analyzed also in [19]. The detailed analyses of the literature data, some critical considerations and the potential ways of optimization of DNA nanopore sequencing were presented in [20]. Some characteristics of ISFET and EIS based DNA sensors are analyzed detailed by us in [21] [22].
In this paper a modified architecture/design for measuring cell of DNA sequencing using tunneling current is offered in order to control and optimize the translocation speed of the DNA molecules. We study the features of the dynamics of the movement of DNA molecule in an aqueous solution under gravity, electrophoretic and drag forces in order to reduce the rate of movement of the DNA and increase DNA reading time.
2. DNA Dynamics and DNA Speed in Aqueous Solution
In Figure 1 the scheme of architecture/design for investigation of DNA nucleosides sequencing by the solid state nanopore modified method is presented. The electrolytic cell filled by aqueous solution provides with nanopore, reference and back electrodes (RE and BE). The electrical potential applied on the RE/BE promotes directional movement of the charged DNA molecules under the action of an electric field. The potential applied to the lateral metal electrodes M1 and M2 facilitates the creation of molecular junctions with nucleosides. A nanopore consisting of gold electrodes is covered by insulator layer. Contacts to nanopore can be made in the form of nanowires or nano-ribbons (Figure 1(c)). Contacts surface area coated by the insulator layer for its protection and for not letting the negatively charged molecules stick the surface of the electrodes. Capture of DNA molecules on the gold electrode surface will create “false” currents and distort the useful signal. If necessary, many nanopores can be mounted in a cell and a multi-nanopore chamber can be created and at the same time many DNA molecules can be studied (Figure 1(b)). In Figure 1(d) example of molecular junction M1-DNA GC base pair-M2 is presented.
At the bare pore under applied voltage V between metallic electrodes M1 and M2 flow only low ionic current of electrolyte
in order to pA [6] [23] [24]. Ionic current through the channel sharply decreases (is blockade) while the DNA molecule move under the gravity
, electrophoretic force
(migration under applied potential on the reference electrode), drag force
and diffusion (connected with ions concentration gradient in solution) become to the pore and blockade the ionic current. When some of nucleotides of DNA [adenine (A), guanine (G), thymine (T), cytosine (C), nucleotide pairs GC, AT, etc.] captured between gold electrodes, they create the electronic bridge and consequently molecular junctions (at the
, see [6]). So, DNA bases can create individual electronic bridges between gold electrodes and across a pore (through the nucleotide junction) that will flow only tunnel current. Metal electrodes can be made in the form of nanowires/nanoribbons to create, if possible, a large area of coverage of the coming close DNA molecules with several orientations (Figure 1(c)). Such a design of the electrodes can be realized using mechanically controllable break-junction (MCBJ) [25] [26] [27], scanning tunnel microscope (STM) [28] or atomic force microscope (AFM) [29] techniques.
![]()
Figure 1. (a) The measurement cell of investigation of DNA nucleosides by the solid state nanopore modified method. (b) Multi-nanopore (4 nanopore) design. (c) Nanogape with nanowire or nanoribbon gold electrodes. (d) Example of molecular junction M1-DNA GC base pair-M2. The potential applied to the electrodes [Au(111)] facilitates the creation of a molecular junction with nucleosides. The Au(111) substrate was chosen as a noble inert substrate to minimize molecule-substrate interactions. Vertical ionic current
depend with potential
, applied to RE. Lateral ionic current
conditioned by potential applied to metallic electrodes. RE means reference electrode, BE means back electrode. It is showing also the gravity (
), electrophoretic (
) and drag (
) forces, influences on molecules and controlled their movement.
Such a design of electrodes will contribute to almost ~100 percent creation of molecular junctions at any orientation of the DNA. The gap between electrodes must be so narrow that only one DNA molecule passes through it.
For effective reading of nucleotides, it is necessary that they move vertically. The vertically directional movement of the molecule will be determined by the resulting force
(1)
Let’s assume that distribution of the DNA molecules in aqueous solution is homogeneous and DNA concentration gradient in the solution is absence or very small. Then ignoring the role of diffusion and considering that
,
,
, (2)
for the magnitude of the resulting force we can write:
. (3)
Here
is the mass of DNA molecule,
is the applied voltage on the reference electrode,
is the distance between RE and nanogap,
is the electric field acting on DNA, g = 9.80665 m/s2, q is the negative charge of DNA,
is the speed of the molecule relative to the aqueous solution, A is the molecule cross sectional area, 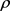 is the density of the solution1, and
is the density of the solution1, and 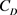 is the drag coefficient—a dimensionless number2.
is the drag coefficient—a dimensionless number2.
Let’s discuss the magnitudes of
,
and
.
For definiteness assume that DNA consists only from
blocks of A, G, C and T nucleotides. Then for
we have:
.
Here 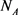 is the Avogadro number,
is the Avogadro number, 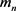 is the mass of group from four nucleotides,
is the mass of group from four nucleotides, 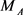 ,
,  ,
, 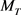 and
and 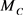 are the molecular masses of adenine (C5H5N5), guanine (CH5N5O), thymine (C5H6N2O2) and cytosine (C4H5N3O), correspondingly. For example, at the
are the molecular masses of adenine (C5H5N5), guanine (CH5N5O), thymine (C5H6N2O2) and cytosine (C4H5N3O), correspondingly. For example, at the , we have
, we have 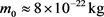 and
and
 .(4)
.(4)
The value of 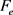 depends on
depends on 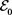 and for example at the single charged DNA (
and for example at the single charged DNA (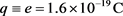 ),
),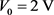 and at the
and at the 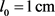 for the
for the 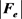 we have:
we have:
![]() .(5)
.(5)
Lets’ consider the value![]() . Taking
. Taking ![]() (as a water,
(as a water,
pH = 7), ![]() [9],
[9], ![]() (
(![]() is the DNA
is the DNA
diameter [31]) we receive
![]() .(6)
.(6)
Where molecule speed ![]() given in m/s.
given in m/s.
Now let’s determine functional dependency of molecule translocation speed ![]() vs. other parameters, especially vs.
vs. other parameters, especially vs. ![]() and
and![]() . Molecule move under resulting force (3). Kinetic energy of the molecule is equal to work of resulting force. Taking the aqueous solution surface as the origin of coordinate and assume that initial coordinate of molecule is the
. Molecule move under resulting force (3). Kinetic energy of the molecule is equal to work of resulting force. Taking the aqueous solution surface as the origin of coordinate and assume that initial coordinate of molecule is the ![]() (Figure 1) and DNA initial speed is
(Figure 1) and DNA initial speed is ![]() we can write
we can write
![]()
Then on the distance ![]() molecule will accumulate the kinetic energy equal to work of resulting force and
molecule will accumulate the kinetic energy equal to work of resulting force and
![]() ,(7)
,(7)
оr
![]() ,
,
аnd
![]() .(8)
.(8)
One of significant problems of DNA sequencing is to decrease translocation speed. Assume that molecule starts moving from upper side of chamber (![]() ) with zero starting speed (
) with zero starting speed (![]() ). For regulating the DNA speed as variable parameters, we can choose
). For regulating the DNA speed as variable parameters, we can choose ![]() and
and![]() . It is clear also that
. It is clear also that ![]() and for minimizing
and for minimizing ![]() we must put “-” sign in (8) before
we must put “-” sign in (8) before ![]() and consider the condition
and consider the condition
![]() or
or ![]() .(9)
.(9)
Then expression (8) we can rewrite as
![]() .(10)
.(10)
At the ![]() (
(![]() ),
),![]() and
and ![]() the condition (9) equivalent to:
the condition (9) equivalent to:
![]() .
.
Measurements of blockade current of ssDNA in [32] imply that, while polymers longer than the pore they are translocated at a constant speed, the velocity of shorter polymers increases with decreasing length. Note that unlike our case in [32] polarity of the potential applied to gate electrode and electric force directed to opposite (Figure 1(a) in [32]), and velocity quadratically depends on applied field.
As we can see the magnitude of DNA translocation speed strongly depend on ![]() and the DNA size, particularly on
and the DNA size, particularly on![]() . Using the above parameters, we obtain
. Using the above parameters, we obtain
![]() .
.
For the “optimal” case ![]() from (10) we have
from (10) we have
![]() .
.
The schematic behaviors of the molecule speed vs. ![]() and
and ![]() are shown in Figure 2. DNA translocation speed dramatically decreases with the electric field. About such behavior of
are shown in Figure 2. DNA translocation speed dramatically decreases with the electric field. About such behavior of ![]() is noted also in [10] [11]. Note that the calculated results of [11] predict that for long polymers the translocation mean velocity, defined as the ratio of the polymer contour length and the average first passage time, approaches a constant value that does not depend on
is noted also in [10] [11]. Note that the calculated results of [11] predict that for long polymers the translocation mean velocity, defined as the ratio of the polymer contour length and the average first passage time, approaches a constant value that does not depend on![]() . This type of dependence is consistent with our dependencies at the higher values of
. This type of dependence is consistent with our dependencies at the higher values of ![]() (saturation of
(saturation of![]() ) (Figure 2(b)).
) (Figure 2(b)).
For receiving low value of speed, we need to minimalized term (![]() ) by the varying
) by the varying ![]() and
and![]() . Problem is to find optimal values of
. Problem is to find optimal values of![]() .
.
Let’s introduce dimensionless electric field strength ![]() and dimensionless speed
and dimensionless speed ![]() as follows:
as follows:
![]() ,
,![]() .(11)
.(11)
Now expression (10) we can rewrite as follows:
![]() , or
, or ![]() .(12)
.(12)
For receive low speed it is necessary that ![]() will be minimal, or with values of
will be minimal, or with values of ![]() very close to unity.
very close to unity.
Numerical calculations of the DNA translocation speed vs. ![]() are carried out. Results of numerical analyzes for DNA three sizes (
are carried out. Results of numerical analyzes for DNA three sizes (![]() ,
,![]() and
and![]() ) presented in Table 1. Low magnitude of
) presented in Table 1. Low magnitude of ![]() is obtained with
is obtained with
![]() (a) (b)
(a) (b)
Figure 2. Schematic dependency of the molecule speed vs. ![]() (a) and
(a) and ![]() (b).
(b).
low values of ![]() and for short DNA (low values of
and for short DNA (low values of![]() ). For comparison in Table 2 is presented some literature data of DNA translocation speed.
). For comparison in Table 2 is presented some literature data of DNA translocation speed.
The comparison of the results of Table 1 and Table 2 shows the advantage of the proposed method of reducing the translocation speed. Speed control is achieved through the accurate compensation of gravity and drag forces. This can be achieved by controlling the potential of the reference electrode. By adjusting the size of DNA molecule and increasing the accuracy of changing of the reference electrode potential, the translocation speed can be significantly reduced (see cases ![]() or 10).
or 10).
In Figure 3 the logarithmic dependency of dimensionless DNA translocation speed vs. dimensionless field ![]() is presented for the case
is presented for the case ![]() according to data of Table 1. This graph built for the case of the optimal values of
according to data of Table 1. This graph built for the case of the optimal values of ![]() and
and![]() . In this linear dependency it also shows corresponding values of DNA molecule translocation speed in nm/s units for several values of
. In this linear dependency it also shows corresponding values of DNA molecule translocation speed in nm/s units for several values of![]() .
.
The main difficulty of such way to decrease translocation speed conditioned
![]()
Table 2. Literature data of DNA translocation speed.
![]()
Figure 3. Logarithmic dependency of DNA translocation dimensionless speed vs. dimensionless electric field. It is shown values of molecules speed in nm/s for the case ![]() (the data on the right site) and
(the data on the right site) and ![]() (the data on the left site).
(the data on the left site).
by the very slightly changing of electric field![]() . The translocation velocity varies also depend on parameters such as the electrical potential, the type of nanopore, and whether the DNA is single-stranded or double-stranded (see also [34]).
. The translocation velocity varies also depend on parameters such as the electrical potential, the type of nanopore, and whether the DNA is single-stranded or double-stranded (see also [34]).
Elevator Method
An alternative method for reducing DNA translocation time can be the so-called “elevator method”. By the elevator method, we mean the situation when the nanopore construction (metallic electrodes) moves parallel to Z axis along with the DNA (Figure 1). Moreover, the speed of movement of a nanopore must be approximately equal to or slightly weaker than the speed of movement of DNA. The “elevator method” can be easily implemented in Space (in the Shuttle) without gravity. At the same time, the depth of the cell with an aqueous solution must be quite large so that during the time of passage of the nanopore, at least several DNA molecules can pass through the pore. In this case, the time of passage of the molecule around the electrodes will increase, and the time it takes to read the molecule will be relatively long. The movement of the electrodes can only be adjusted mechanically. Regulation of the pore movement speed by electromagnetic forces is probably impossible, because firstly the electrodes themselves are charged and secondly, they must move in the field created by the potential![]() .
.
3. DNA Molecules Reading (Translocation) Time
Time for DNA molecules reading (translocation) can be set by the duration of the electric pulse on the metal electrodes (see [6]). Denote this time by![]() .
.
By the sizes of gap between nucleotides ![]() (see [35]) we can determine the time of absence of the applied impulses on the metallic electrodes [6]. This time can be calculated according to formula
(see [35]) we can determine the time of absence of the applied impulses on the metallic electrodes [6]. This time can be calculated according to formula![]() , or
, or
![]() ,(13)
,(13)
The period of the pulses applied to the gold electrodes will be
![]() .(14)
.(14)
The frequency of the base’s passes (i.e. electrical pulses applied to lateral gold electrodes) will be
![]() .(15)
.(15)
Here ![]() is the time for DNA molecules bases (A, T, C or G) reading (translocation), and
is the time for DNA molecules bases (A, T, C or G) reading (translocation), and
![]() ,(16)
,(16)
![]() is the vertical size of corresponding nucleotides (
is the vertical size of corresponding nucleotides (![]()
![]() [see Appendix]),
[see Appendix]), ![]() is the size of gap between nucleosides [35].
is the size of gap between nucleosides [35].
Example 1: ![]() and
and![]() ,
,![]() ;
;![]() , and
, and![]() , we get
, we get![]() ,
,![]() ;
;![]() , and
, and![]() , we get
, we get![]() ,
,![]() .
.
Example 2: ![]() and
and![]() ,
,![]() ;
;![]() , and
, and![]() , we get
, we get![]() ;
;![]() ;
;![]() , and
, and![]() , we get
, we get![]() ;
;![]() .
.
If t is the passing full time for one DNA molecule through nanopore and ![]() is the number of bases groups in DNA, we can write
is the number of bases groups in DNA, we can write
![]() ,(17)
,(17)
where
![]() (18)
(18)
is the DNA total length consisting only groups with four bases. Here ![]()
![]() ,
, ![]() ,
, ![]() and
and ![]() are the sizes in direction of Z axis of adenine, thymine, guanine and cytosine, correspondingly (see Appendix).
are the sizes in direction of Z axis of adenine, thymine, guanine and cytosine, correspondingly (see Appendix).
In the other hand
![]() .(19)
.(19)
For example, for the case![]() ,
, ![]() and
and![]() , we get
, we get![]() .
.
It is quite large and measurable time for reading of DNA sequencing.
We can control value of ![]() and f by the choice magnitudes of
and f by the choice magnitudes of ![]() and
and![]() . For comparison note that in Ref. [7] the reasonable value of the DNA translocation speed is near to 0.01 - 1 ms per base considered. It is more short time compared with 0.031 s. Note that all of the biological and synthetic nanopores have barrels of ∼5 nm (which is considerably longer than the base-to-base distance of 3.4 Å) in thickness and accommodate ∼10 - 15 nucleotides at a time. It is, therefore, impossible to achieve single-base resolution using blockage current measurements. In addition, the average rate at which a polymer typically translocates through a nanopore is on the order of 1 nucleotide/μs (i.e., on the order of MHz detection), which is too fast to resolve.
. For comparison note that in Ref. [7] the reasonable value of the DNA translocation speed is near to 0.01 - 1 ms per base considered. It is more short time compared with 0.031 s. Note that all of the biological and synthetic nanopores have barrels of ∼5 nm (which is considerably longer than the base-to-base distance of 3.4 Å) in thickness and accommodate ∼10 - 15 nucleotides at a time. It is, therefore, impossible to achieve single-base resolution using blockage current measurements. In addition, the average rate at which a polymer typically translocates through a nanopore is on the order of 1 nucleotide/μs (i.e., on the order of MHz detection), which is too fast to resolve.
4. Conclusions
Solid-state nanopore DNA sequencing modified method is developed and presented. The nanogap is made on gold electrodes in the form of nanowires or nanoribbons. A detailed analysis of the dynamics of a DNA molecule in an aqueous solution is carried out. For the first time in analytical calculations, all forces acting on DNA molecules are taken into account. The movement of DNA in aqueous solution is regulated by the potential applied to reference electrode. Taking into consideration that DNA moves under gravity, electrophoretic and drag forces of the expression for the DNA translocation speed are calculated and analyzed. The conditions for decreasing the DNA translocation speed or increasing reading time are received. Carried numerical calculations show that increasing the reading time is associated with the problem of very subtle regulation of the electric field strength acting on DNA molecule.
Based on the modified method proposed above it is possible to successfully decrease molecule translocation time and increase its reading time. It will give possibility of enlarging the passing time of tunnel current through gold-nucleotide-gold junction and more precisely determine and identify nucleotide type crating an electrical bridge between electrodes [6]. Our results therefore potentially suggest a realistic, inherently design-specific, high-throughput nanopore DNA sequencing device/cell as a de-novo alternative to the existing methods.
The amount of tunnel current which can pass through the nanopore at any given moment therefore varies depending on whether the nanopore is bounded by an A, T, C or G nucleotide. The change in the current through the nanopore as the DNA molecule passes through the nanopore represents a direct reading of the DNA sequence. Such behavior is ordinary. Analyses of the tunnel currents responsible for nitrogenous bases in DNA are developed and presented by us in [6].
It has been shown that one can control value of the DNA molecules bases reading time and the frequency of the electrical pulses applied to lateral electrodes by the choice of magnitude of the potential of the reference electrode. It is shown that DNA reading time can be quite large and measurable value in order to 103 bases/s. The conditions of lengthening of the reading time are determined.
Author Contribution Information
All authors participated in the statement of the problem and discussion of the results. L. Gasparyan, I. Mazo and F. Gasparyan conducted literature review. F. Gasparyan and V. Simonyan made calculations and participated in the writing of the text of the article.
Appendix
For calculations of nucleosides sizes, we use data for the covalent radii some elements presented in TableA1 [36].
![]()
Table A1. The covalent radii some elements in nm [36].
Let's denote the vertical size of the nucleotides through h, and the horizontal size—through l. Indexes H-C (or C-H), C=N etc. means single and double bonds between corresponding elements.
Adenine (C5H5N5)
![]()
![]()
![]()
Guanine (CH5N5O)
![]()
![]()
![]()
Thymine (C5H6N2O2)
![]()
![]()
![]()
Cytosine (C4H5N3O)
![]()
![]()
![]()
NOTES
1Density of the fluid determined by the expression
[30], where
is the percent composition. Density of the fluid changed very little, e.g. at the
,
.
2The drag coefficient
depends on the shape of the object and on the Reynolds number
. Here D is some characteristic diameter or linear dimension and
is the kinematic viscosity of the fluid. At low
is asymptotically proportional to
, which means that the drag is linearly proportional to the speed. At high
is more or less constant and drag will vary as the square of the speed. In the case of high velocity, in general,
a function of the orientation of the flow with respect to the molecule (apart from symmetrical objects like a sphere). Assuming that surface area of the DNA molecule end is the half-sphere in further calculations we can take
[9].