Received 3 March 2016; accepted 8 May 2016; published 13 May 2016

1. Introduction
According to the World Health Organization, probiotics are “live microorganisms which, when administered in adequate amounts, confer a health benefit on the host” [1] . Probiotic strains with the ability to produce antimicrobial compounds are often used to control the growth of pathogenic microorganisms in fermented food and animal feed. The strains may also be incorporated into personal care products [2] [3] . Daily supplementation with probiotics proofed effective in the alleviation of cold and flu symptoms [4] , and as food supplement to patients on cancer treatment [5] . Many probiotic bacteria produce a broad range of effective antimicrobials, including lactic acid, hydrogen peroxide and bacteriocins.
Bacteriocins are commonly defined as genetically encoded substances of a proteinaceous nature produced by virtually all bacteria and are active against various, most closely-related, microorganisms [6] . This positions them as a very appealing alternative to antibiotics and chemical stressors, especially in an age when alternative bacterial infection therapies are being widely investigated [7] .
Several strains of Bacillus spp. have been recognized as safe for food or industrial applications and, importantly, have been documented as probiotics [8] .
This study investigates several characteristics of Bacillus subtilis KATMIRA1933 and Bacillus amyloliquefaciens B-1895, both of which are suspected to have probiotic properties. B. subtilis KATMIRA1933, isolated from a dairy product called Yogu Farm, has been unwittingly consumed by humans for years without harmful effects [9] . The strain produces antimicrobial proteins, including subtilosin A [9] . Genomic analysis has shown that B. amyloliquefaciens B-1895, isolated from soil, has the potential to produce a number of proteolytic enzymes and subtilin, an antimicrobial peptide active against foodborne pathogen Listeria monocytogenes [10] . The strain is used as a probiotic in royal fish [11] . Inclusion of B. amyloliquefaciens B-1895 into bird feed enhanced immunity to the Newcastle virus [12] and improved the body weight of the birds [13] . In addition, fermentates of both strains were reported as having antioxidant and DNA protective activities [14] .
Any microorganism considered for use as a probiotic must be tested for specific advantageous characteristics, balanced by a thorough evaluation of its safety. The antibiotic susceptibility of bacterial cells to routinely prescribed antibiotics is essential for the putative probiotic’s safety evaluation. Microbial resistance to antibiotics in part is a gene-encoded mechanism that occurs either by genetic mutation or by gaining resistance via horizontal or vertical gene transfer [15] . It is important to find out if antibiotic-resistance gene(s) are transferable from probiotics to the commensal microorganisms or to pathogenic bacteria. New forms of resistant pathogens may emerge if such genes are transferred from probiotics to pathogens [16] . Many studies have been conducted to identify the antibiotic-resistance genes in Bacillus species [17] - [20] .
Some products produced by bacteria have the potential to damage host cells. It is thus important to screen for such products when assessing the safety of a strain. In particular, hemolysin production by bacteria has been identified as a virulence-associated feature [21] . Various bacterial species, including Bacillus cereus and group A streptococci, which produce hemolysin BL and streptolysin-o, respectively, are considered as pathogens due to their potent hemolytic activity [22] . The hemolytic mechanism in Bacillus is not fully understood. However, recent studies have been performed to identify the gene(s) responsible for hemolytic activity [23] [24] . Even though the hemolytic activity of some Bacillus spp., such as Bacillus subtilis, is less than in pathogens [25] , these isolates may be considered unsafe for food or personal health care applications until the effect of this virulence factor is either eliminated, modified, or confirmed as causing no harm to the eukaryotic host.
Bacillus species have a long history of use in biotechnology and as dietary supplements for humans and animals of agricultural importance. Also, bacilli have been engineered to produce biologically active substances such as antibiotics and enzymes [26] . A distinctive feature of the Bacillus spp. is high proteolytic activity [27] . The advantages and importance of proteolytic enzymes have been widely reported. Briefly, they include the activation of regeneration processes, the enhancement of normal digestion, and degradation of allergic compounds [27] - [29] .
Mutagenicity and carcinogenicity assessments of the antimicrobial substances are the harbinger to efficiently evaluate bacterial products possessing antibacterial activity prior to their use in pharmaceutical applications. The Ames test is used to evaluate the mutagenic potential of substances by determining if the chemical causes DNA damage that leads to genetic mutations in Salmonella spp. [30] . Association between mutagens and cancer induction [31] raises the importance of recruiting a screening test for mutagenicity to ensure the safe consumption of such chemicals.
As many probiotics are to be consumed, their selection depends on bacterial tolerance to acids and bile salts. Candidacy of a probiotic depends on the ability of bacterial cells or their spores to survive and grow at the high acidity (pH 3 or below) of the stomach [32] and with the detergent-like activity of intestinal bile salts that disrupt the cellular membrane [33] . In vitro, the tolerance of Bacillus species to acids and bile salts reflects their survival rates and viability through the gastrointestinal tract [32] [34] .
Coaggregation is the adherence of genetically distinct bacteria and is considered as a desirable characteristic in a probiotic microorganism [35] . It is believed to facilitate the integration of exogenous bacteria, which is important for the development of multispecies biofilms [35] . Coaggregation allows a beneficial organism to adhere to a pathogenic organism. Aggregation may also help the bacterium adhere to different surfaces, which is very relevant to human health. Adhesion may also allow probiotic organisms to create a barrier, which could effectively prevent colonization by pathogens [36] . Auto-aggregation of probiotic strains may be required for adhesion to intestinal epithelial cells, which would keep them from being flushed out by the body, and would in such a way give them an advantage over other organisms [36] . Though not all mechanisms of action have been determined, further research can reveal the more complex aspects and implications of this ability.
The objective of this study is to investigate whether B. amyloliquefaciens B-1895 and B. subtilis KATMIRA 1933 possess additional health benefits that would qualify them to be probiotic candidates. A battery of tests is commonly employed and has been addressed in the study on B. subtilis KATMIRA1933 and B. amyloliquefaciens B-1895. Antibiotic susceptibility, hemolytic/fibrinolytic activity, proteolytic activity, bacterial reverse mutation, tolerance to acids and bile salts, and bacterial auto-aggregate/co-aggregate abilities were evaluated and analyzed for both bacilli. The antimicrobial activity of Cell-Free Supernatants (CFSs) of tested bacilli against pathogens was also evaluated. Regarding the ability to auto-aggregate and co-aggregate, the most appropriate method for the mathematical interpretation of collected data was identified. A visual analysis utilizing microscopy was used as a mode of comparison for evaluating two methods of mathematical analysis. It was determined that both the extracts possessed unique antibacterial capabilities along with the desirable traits that would allow them to progress as candidates for probiotics therapies.
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
Frozen stocks of B. subtilis KATMIRA1933 and B. amyloliquefaciens B-1895 were inoculated in De Man, Rogosa and Sharpe (MRS) medium (Becton Dickinson and Company, Sparks, Maryland, USA) and incubated aerobically with shaking at 250 rpm for 24 h at 37˚C. M. luteus ATCC10240 was used as a reference microorganism. The pathogenic bacteria included in our study are listed in Table 1. These bacterial strains were inoculated into tryptic soy medium (Becton Dickinson and Company, Sparks, Maryland, USA) and incubated aerobically for 24 h at 37˚C. The oral pathogens, upon revival from −80˚C DMSO stocks, were maintained on trypticase soy broth (TSB) and agar plates containing haemin 1 µg・mL−1, menadione 1 µg・mL−1, 20 % defibrinated sheep’s blood (BAPHK) at 37˚C under anaerobic conditions (5% H2, 10% CO2, 85% N2). S. intermedius strain F0413 was maintained on Brain Heart Infusion broth at 37˚C under anaerobic conditions, while S. mutans strain 25,175 was incubated aerobically. Broth cultures of Porphyromonas gingivalis strains, Prevotella intermedia strains, and Fusobacterium nucleatum were grown in Todd?Hewitt broth (THB) containing haemin 1 µg・mL−1 and menadione 1 µg・mL−1 (designated THBHK) at 37˚C under anaerobic conditions.
2.2. Antimicrobial Activity of CFS of Studied Bacillus Strains
Cell free supernatant of two tested bacilli strains were prepared as previously described by Sutyak et al. [9] . Broth cultures (1.5 mL) of oral pathogens were grown for 24 h in the appropriate atmosphere and media. Culture density for each strain was determined using OD600 values. All strains were adjusted to an OD600 of 0.1 to correspond to 1 × 106 CFU・mL−1. Cultures were swabbed to the appropriate solid support agar media, which had
![]()
Table 1. Bacterial strains and culture conditions used in this study.
aCNS: Central Nervous system, bGI: Gastrointestinal.
been dried for 20 min in a tissue culture hood. Plates that were to be used under anaerobic conditions were placed in the chamber and all further steps were carried out within. Using the wide end of a 200 µL yellow pipette tip, holes were bored into the media, creating a well to accommodate supernatants. Cell-free supernatant (120 µL) from B. subtilis KATMIRA1933 and B-1895 were added to each well, in triplicate. Plates were incubated for 5 days at 37˚C under anaerobic conditions and the diameter of the zones of inhibition were measured in millimeters (mm) with a digital caliper.
2.3. Antibiotic Susceptibility of Studied Bacilli
B. subtilis KATMIRA1933, B. amyloliquefaciens B-1895, and M. luteus ATCC 10,240 were included in this assay. Bacterial strains grown overnight were diluted 1:100 with corresponding fresh media to yield approximately 106 CFU・mL−1. This was verified by the plate counting method. The disc diffusion test was conducted according to the (CLSI) Performance Standards for Antimicrobial Disk Susceptibility Tests [37] . The tested antimicrobial discs included ampicillin (10 µg), erythromycin (15 µg), and tetracycline (30 µg) from Becton Dickinson and Company (Sparks, Maryland, USA), while bacitracin (10 µg), chloramphenicol (30 µg), kanamycin (30 µg), penicillin (10 IU), streptomycin (10 µg) and oxytetracycline (30 µg) were from Benex Limited (Shannon, Co., Clare). Plates were incubated for 24 h at 37˚C. Radii of zones of inhibition were measured in millimeters (mm) with a digital caliper (Fischer Scientific, Pittsburg, PA, USA) from the edge of the disk to the edge of the inhibition zone.
2.4. Bacterial Reverse Mutation (Ames) Assay
The mutagenicity assessment of subtilosin, CFSs of B. subtilis KATMIRA1933 and B. amyloliquefaciens B-1895 was carried out according to Maron and Ames [38] , Cappuccino and Sherman and [39] with minor modifications. Briefly, Salmonella typhimurium K-6 TA1535 overnight growth was prepared according to the manual a day before performing experiment. Top agars were melted using a hot water bath (45˚C). Then, 300 µL of histidine/biotin and 100 µL of the overnight culture of S. typhimurium K-6 TA1535 were added to the top agars. Top agar contents were gently mixed and immediately poured over the minimal agar plates. A sterile forceps was used to pick up a filter paper disc and dip it into the two microcentrifuge tubes containing different concentrations of tested samples. Positive and negative controls were included. After filter paper discs were saturated with tested chemicals, they were placed into the center of a minimal agar plate that was laid out with top agar containing biotin and bacterial growth suspension. All the plates were incubated aerobically at 37˚C for 48 - 72 h in an inverted position. The results were evaluated by counting the number of colonies that grew on the agar plates. The mutant frequency was calculated for tested samples. Mutant frequency was calculated as the number of revertant colonies in treated plates divided by their numbers in the negative control.
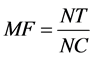
where MF = Mutant Frequency, NT = Number of revertant colonies on treated plates, NC = Numbers of colonies on negative control plate.
The results were expressed as following: the substance considered has mutagenic activity when its MF value is ≥2, is a possible mutagen when the value ranges of 1.7 to 1.9, and has no mutagenic activity when the frequency ranges ≤ 1 - 1.6 [40] .
2.5. Determination of Protease Activity by the “Stabbing” Method
Detection of the proteolytic activity of Bacillus strains, B. subtilis KATMIRA1933 and B. amyloliquefaciens B-1895, was performed as described by Ponmurugan [41] with minor modifications. Briefly, from the frozen stocks, bacilli were maintained in MRS broth aerobically with agitation (150 rpm), for 24 h at 37˚C. From the last overnight culture, 10 µL was spread on MRS agar by streaking with a loop and plates were incubated aerobically for 24 h at 37˚C. After the incubation period, one colony was picked up using an inoculating loop and stabbed into a milk agar plate. A milk agar composed of peptone (0.1%), NaCl (0.5%) and skim milk (10%) was prepared according to Uyar et al. [42] with some minor modifications. The components of the milk agar medium were mixed thoroughly with double deionized water and autoclaved for 15 min. The inoculated plates were incubated for 24 h at 37˚C under aerobic conditions. The results were reported as following: a clear zone of proteolytic activity around inoculated colonies represented protease positive and if the clear zone did not appear, it was protease negative.
2.6. Hemolytic Activity on Whole and Defibrinated Blood Agar
A hemolytic assay was performed as described by Luo et al. [43] with minor modifications. Instead of inoculating blood agar with 10 µL of bacterial suspension using a disposable loop, a polyester tipped applicator (Fischer scientific, Pittsburg, PA) was used to spot-inoculate onto the whole and defibrinated blood agar. This was achieved by touching the tip of the applicator to one bacterial colony before using the tip to lightly touch the fresh blood agar while rotating the applicator. By using the polyester applicators, circular inoculation sites of about 5 mm in diameter were formed. Each bacterial strain was inoculated onto the blood agar using this method, and sufficient space was given between each spot. Inoculated blood agar plates were then incubated aerobically for 24 h at 37˚C. Plates were then checked for hemolytic activity.
2.7. Fibrinolytic Activity Test
One milliliter of LB broth was inoculated with B. amyloliquefaciens B-1895 and B. subtilis KATMIRA1933 and incubated for 18 h at 37˚C with agitation. Each culture was adjusted to one unit of the MacFarland Turbidity standard. Then, 1 mL of the adjusted cell suspensions and 24 mL of fresh LB were mixed in 50 mL culture flasks, and incubated 5 days at 37˚C. The CFSs were collected by centrifugation (4480 g, 15 min, and 4˚C). Plates containing fibrin were prepared accordingly, the 1.5% agar-based formulation (10 mL) consisting of 0.4% fibrinogen in 50 mM sodium phosphate buffer at pH 7.4 was mixed with 0.1 mL of thrombin (10 NIH units) poured into the Petri dishes (15 mL), and left to polymerize and to dry at room temperature. Then, 7 mm diameter holes were punched in the solidified agar. These holes were filled with 30 μL of the CFSs and incubated overnight. The clear zones of fibrinolytic activity were measured with a digital caliper.
2.8. Coaggregation Test
The coaggregation assay was performed to evaluate the coaggregation ability of B. subtilis KATMIRA1933 and B. amyloliquefaciens B-1895 strains with select pathogens following the method described by Cisar et al. [44] with some modifications. Bacterial cultures were harvested from the planktonically grown cells incubated at 37˚C bycentrifugation (4480 g, 15 min, 23˚C); they were washed with sterile phosphate buffered saline (PBS) twice. After the second wash, the harvested cells were re-suspended in PBS and the optical density (OD600) was adjusted to 0.25. In a 96-well microtiter plate, 100 µL of each test strain was mixed with 100 µL of Bacillus strain, while 200 µL of each bacterial suspension in monoculture was used as controls. The plate was placed in a micro-titer plate spectrophotometric reader (SmartSpecTM 3000) and kept at 30˚C. Measurements of OD600 were taken once per hour for 24 h and calculated for coaggregation. Each experiment was performed in quadruplicate. Samples of 100 µL were taken after 2 h reading for Gram staining and observed microscopically for coaggregation (Figure 1).
2.9. Mathematical Analyses
Two methods were employed to calculate the percent of coaggregation after 2 h incubation in each of the mixtures, for the purpose of comparison. In the first method, the percent auto-aggregation of each bacterium and the percent coaggregation in each mixture was calculated as described by Ledder et al. [35] with the following equation:
Method 1:
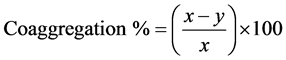
where x is the pre-incubation value and y is the post-incubation value at a certain time point.
The second method employed the equation described by Handley et al. [45] :
Method 2:

where Ax and Ay are the organisms as controls and (x + y) is a mixture of the two.
Both methods used optical density data obtained after 2 h, at OD600. The data from the two methods were compared and the more appropriate method was chosen from the analysis of 2 h and used for the 8 h analysis.
2.10. Microscopy
Bacterial interactions at the 2 h time points were visualized on slides using histological techniques. To visualize, bacteria were stained with Gram stain (BD, Becton and Company, Maryland, US). Images were obtained with a Nikon DS-Fi1 camera mounted on a Nikon Eclipse 80i compound microscope using the 100×/1.25 oil objective. Images were analyzed using Nikon, NIS- Elements D3.0 software. The amount of coaggregation was visually analyzed and scored with a scoring system, with 0 being the absence of coaggregation and 4 being an abundance of coaggregation (Figure 1).
2.11. Production of Bacillus Spores
Sporulation-inducing esculin agar was composed of esculin hydrate (E8250-5G) (Sigma-Aldrich), 1 g, ferric
![]()
Figure 1. Microscopic analysis of auto-aggregation and coaggregation. A 0 - 4 scoring system was utilized, with 0 representing no adhesion between similar or different microorganisms, and 4 repres- enting maximum aggregation after 2 h incubation.
citrate (Fisher scientific), 0.5 g, and BHI, 40 g. These components were mixed and completely dissolved in deionized water up to 1 L. The pH of the mixture was adjusted to 7.0, agar (1.5%) was added and the medium then was autoclaved. Bacillus sporulation was achieved following Franklin and Clark [46] with minor modifications. Briefly, the esculin agar medium was poured into 150 × 15 mm Petri dishes to achieve about a 10 mm thick layer. Bacillus grown on MRS plate was scraped and seeded onto an esculin plate. The inoculated agar was incubated for 15 - 25 days aerobically at 37˚C. After the first five days of incubation, spore production by the Bacillus strains was monitored daily using light microscope. Once sufficient numbers of spores were produced in the grown colonies, they were harvested using sterile inoculating loops. The spores were washed with sterile distilled water and pelleted by centrifugation (5444.5 g, 20 min at 4˚C). The pellets were re-suspended with 20 mL sterile distilled water, glass beads were added, and incubated with agitation at 75˚C - 80˚C for 25 min to ensure the killing of the vegetative cells. Following that, tubes were placed on ice for 10 min and then the glass beads were discarded. The suspension was collected by centrifugation (5444.5 g, for 20 min, at 4˚C) and washed three times with ice-cold sterile water. The spores were re-suspended in a minimum volume of sterile ice-cold water and counted by plating. The spore suspension was aliquoted and stored at −80˚C for future use.
2.12. Acid and Bile Tolerance of Bacillus Spores
The acid and bile tolerance method was performed according to Hyronimus et al. [47] with minor modifications. Briefly, a frozen stock of Bacillus spores was diluted with PBS to achieve 108 spores・mL−1. Tubes containing spores were incubated at 80˚C for 20 min with agitation to get rid of remaining vegetative cells. After heat treatment, the tubes were placed on ice for 10 min. Ten milliliters of MRS broth was transferred into sterile tubes and the broth pH was adjusted to different pH values: 2.0, 2.5, 3.0 using 0.1 N HCl (Sigma Aldrich, St. Louis, MO). Ten milliliters of MRS broth containing 0.3% bile salts (Oxoid Ltd., Basingstoke, UK) was prepared and transferred into 50 mL test tubes. Control tubes containing broth medium only without adding acid or bile salts were included in this experiment. A 100 µL of diluted spore suspension contain 5 × 107 Spore Forming Unit per milliliter (SFU・mL−1) was dispensed into each tube (acid, bile, and control tubes). At each time interval, 0, 0.5, 1, 2, and 4 h, the spread-plate method was used to enumerate the numbers of surviving spores after acid and bile treatment on MRS plates. The inoculated plates were incubated aerobically at 37˚C for 24 - 30 h. For each treatment, the survival rates were measured. Survival rate was defined as the percentages of the logarithmic number of SFU at each time point (s) divided by SFU numbers at 0 time point (control). For example, the survival rates after 4 h is ((log10 SFU・mL−1 at 4 h)/(log10 SFU・mL−1 at 0 h)) × 100.
2.13. Statistical Analysis
For the antibiotic susceptibility assay and antimicrobial activity of CFS of studied Bacillus strains, experiments were performed at least three times in triplicate. Co-aggregation and auto-aggregation experiments were conducted three times; the collected values were then analyzed mathematically and visually. Acid and bile tolerance studies were repeated three times in duplicate and the results shown were expressed as mean (%) ± SD. Student’s t-test with two-tailed distribution (Excel, Microsoft Corporation, US) was used to compare the survival rates (%) of the two studied bacilli strains in the 3 sets of pH during the 4 h incubation.
3. Results
3.1. Antimicrobial Activity of CFS of the Studied Bacilli
Selected pathogens were tested for their sensitivity to the CFS of B. subtilis KATMIRA1933 and B. amyloliquefaciens B-1895 strains (Table 2). All P. gingivalis isolates were sensitive to B. subtilis KATMIRA1933 CFS, as indicated by a zone of clearing of the bacteria (Table 2). S. intermedius was susceptible to CFS of B. amyloliquefaciens B-1895 (Table 2). P. intermedia, F. nucleatum and S. mutans were not inhibited by the CFS at the concentrations tested (data not shown).
3.2. Antibiotic Susceptibility of Bacillus Strains
The sensitivity of the Bacillus strains to nine antibiotics was assessed according to CLSI [48] . The antibiotics were ampicillin 10 µg, erythromycin 15 µg, tetracycline 30 µg, bacitracin 10 µg, chloramphenicol 30 µg, kanamycin 30 µg, streptomycin 10 µg, oxytetracycline 30 µg, and penicillin 10 IU. Antibiotic susceptibility test results revealed that B. subtilis KATMIRA1933 was tolerant to bacitracin and streptomycin, and more susceptible to penicillin, ampicillin and chloramphenicol than other tested antibiotics. The tolerance of B. amyloliquefaciens B-1895 strain to bacitracin, streptomycin, tetracycline and oxytetracycline was more than other antibiotics, and its susceptibility to ampicillin and chloramphenicol was the highest. Antibiotic susceptibility of Bacillus strains was compared to that of M. luteus, which is a frequently used Gram-positive reference bacterium (Table 3).
3.3. Hemolytic and Fibrinolytic Activity Test
Hemolytic activity of the Bacillus strains was determined using MRS medium, supplemented by whole blood and MRS, supplemented with defibrinated blood. While B. amyloliquefaciens B-1895 produced weak hemolysis on whole blood agar plates, azone of clearance was observed with B. subtilis KATMIRA1933 (Table 4). On defibrinated blood agar, both Bacillus strains displayed no hemolytic activity. S. aureus and M. luteus were used as positive and negative controls, respectively, for this set of experiments. Because hemolytic activity of B. subtilis KATMIRA1933 was observed on whole blood agar, we hypothesized that the predominant is the fibrinolytic activity. Using a fibrinolytic activity assay, it was demonstrated that B. subtilis KATMIRA1933 CFS produced a fibrinolytic zone of 14 mm diameter, while B. amyloliquefaciens B-1895 CFS formed a zone of 11 mm diameter.
3.4. Proteolytic Activity Test
To determine whether the Bacillus strains possess proteolytic activity, the presence or absence of a zone of clearance around bacterial growth and/or bacterial CFS on milk agar was determined; clearance is indicative of
![]()
Table 2. Bacillus extract-induced zones of inhibition of oral pathogens.
![]()
Table 3. Antibiotic susceptibility test of Bacillus subtilis KATMIRA1933 and Bacillus amyloliquefaciens B-1895.
aPenicillin was 10 IU.
![]()
Table 4. Hemolytic activity of Bacillus strain and their Cell Free Supernatants (CFS).
++ = Complete β-hemolysis, + = Weak hemolysis, − = no hemolysis.
protein hydrolysis. B. subtilis KATMIRA1933 and B. amyloliquefaciens B-1895 cells or CFS showed proteolytic activity with clear zone areas of 13 ± 0.5/5 ± 0.3 mm and 15 ± 0.6/3 ± 0.3 mm, respectively, after 24 h incubation. E. coli O157:H7 and its CFS were used as negative controls (data not shown).
3.5. Bacterial Reversal Mutation (Ames) Assay
To evaluate the mutagenic potential of the Bacillus extracts and purified compounds, with the hope of ensuring these natural products were free of mutagenic factors, the Ames test was conducted using S. typhimurium strain TA1535. In addition to the isolated subtilosin, CFS of the Bacillus strains was tested. The number of revertant colonies was counted on glucose minimal agar and the mutant frequency (MF) was calculated. The number of revertant colonies when MRS broth (negative control) was tested were 15 - 16 CFU per plate (Table 5); similar results were obtained when phosphate buffer saline (PBS; negative control) was tested (18 - 20 CFU per plate) (Table 5). According to Kirkland [40] , a frequency that ranges from ≤ 1 - 1.6 for the tested substances indicates no mutagenic activity. The mutant frequency of 50 µg・mL−1 subtilosin was 1.4 higher when compared with concentration of 100 µg・mL−1 and 550 µg・mL−1 (Table 5). The MF of 100%, 50%, 25% and 12.5% CFS of B.
![]()
Table 5. Mutagenic and carcinogenic assay (Ames test).
subtilis KATMIRA1933 were 0.87, 0.9, 0.7 and 1, respectively. A low mutant frequency was determined for CFS of B. amyloliquefaciens B-1895, 0.56, 0.5, 0.7 and 0.13 when 100%, 50%, 25% and 12.5% respectively, were assessed.
3.6. Auto-Aggregation and Coaggregation of Bacterial Strains
Kinetic measurements of auto-aggregation and coaggregation of the bacilli with pathogenic bacteria was determined during an 8 h time period using an automated micro-titer plate reader to quantitatively evaluate aggregation efficiency. Bacterial strains varied with respect to the time required to observe a high aggregation value. Two methods were used to evaluate the auto-aggregation and coaggregation percentages (Table 6). The highest percentages were obtained using the calculation of method 1, which was previously described by Ledder et al. (2010). Method 1 was chosen in this study as the more convincible method, compared with method 2, after mathematical interpretation of optical density data; the method was similar inits application and more closely matched conclusions made from the microscopic analysis (Figure 1 and Table 6). The percentage of auto- aggregation of B. amyloliquefaciens B-1895 was the highest (89.5%), while the lowest was of S. aureus and S. enterica (14.3% and 15.4%), respectively (Table 7). B. amyloliquefaciens B-1895 strain was highly co-aggrega- tive with E. coli (47.1%), P. aeruginosa (46.9%), S. enterica (43.9%) and L. monocytogenes (41.9%), but it poorly co-aggregated with S. aureus (29.9%). In the case of B. subtilis KATMIRA1933, the high coaggregation percentage was observed with E. coli (50.3%) followed by P. aeruginosa (49.7%), L. monocytogenes (48.2%) and S. enterica (47.4%), while low coaggregation was observed with S. aureus (34%) and S. mutans (31.8%) (Table 8).
3.7. Tolerance of Bacillus Spores and Vegetative Cells to Acids and Bile Salts
To evaluate the acid and bile salt tolerance of the spores and vegetative cells, the bacilli were exposed to a range of acid (pH 2.0, 2.5 and 3.0) and 0.3% bile salts during 4 h. The survival rates (SR) (%) of B. subtilis KATMIRA1933 spores were constant (unchanged) during the incubation periodat all pHs tested. At pH 2.0, the final SR was 98.4% ± 2.3%, and slightly higher in pH 2.5 and 3.0, which were 98.75% ± 1.76% and 99.15% ± 1.2%, respectively. The 4 h SRs for B. amyloliquefaciens B-1895 spores similarly were nearly constant across all pH conditions; in the pH 2.0 and 3.0 environment; the SR was 96.45% ± 5.02%, while at pH 2.5 it was 97.3% ± 3.81%. No significant difference was found in the survival rates of both B. subtilis KATMIRA1933 and B. amyloliquefaciens B-1895 into the three sets of pH values for 4 h. Bile tolerance of the Bacillus spores was 88% ± 1.27% for B. subtilis KATMIRA1933 and 84.85% ± 2.05% for B. amyloliquefaciens B-1895.
![]()
Table 6. Comparison between method 1 (as described by Ledder et al. [35] ) and method 2 (as described by Handley et al. [45] ) to calculate the coaggregation values of Bacillus strains with tested pathogens after 2 h incubation.
![]()
Table 7. Highest auto-aggregation of the tested microorganisms as observed during 8 h of incubation.
![]()
Table 8. The highest co-aggregation % of Bacillus subtilis KATMIRA1933 and Bacillus amyloliquefaciens B-1895 with the selected pathogens during 8 h of incubation.
As expected, the tolerance of vegetative cells of B. subtilis KATMIRA1933 and B. amyloliquefaciens B-1895 strains to acidity was much less than those of the spores. In the case of the B. subtilis KATMIRA1933 strain, vegetative cells were incapable of surviving at pH 2.0 and pH 2.5 after 1 h incubation, however at pH 3.0 the SR was 23.9% at 4 h. B. amyloliquefaciens B-1895 vegetative cells were more tolerant to acidic conditions than those of B. subtilis KATMIRA1933. About 30% of vegetative cells of B. amyloliquefaciens B-1895 were able to survive after 4 h at MRS with the different pH values. There was no difference in the tolerance of vegetative cells of studied Bacillus strains to bile salt when compared with the tolerance of their spores (Data not shown). Statistically significant differences in acid tolerance between the Bacillus strains were observed after 30 min of incubation (Data not shown). While there was not a statistically significant difference between the vegetative cells of both Bacillus strains (P > 0.05), the survival rates (%) in 0.3% bile salts exhibited a statistically significant difference between the spores of the two bacilli strains during the 4 h incubation (P < 0.05) (Data not shown).
4. Discussion
The B. subtilis KATMIRA1933 strain was used as a source of subtilosin A and studied for various applications such as for control of food-borne and other human pathogens [9] . B. amyloliquefaciens B-1895 strain was reported as a putative probiotic in poultry and fish [13] . Based on the beneficial properties of both microorganisms, we hypothesize on their possible use as probiotics for human and/or agricultural applications. Therefore, the effort was made to look into these strains’ safety and some of their probiotic-related capacities.
Positive health effects were noticed such as increment food consumption and increasing of broilers’ body weight [13] . An association between Bacillus-food supplements and immune system stimulation was identified in chicken. When birds were vaccinated against the Newcastle virus, the antibody titers in chicken with a B. subtilis-direct feed were greater than those in the control group without bacilli supplement-food [12] .
The soil isolate, B. amyloliquefaciens B-1895 showed probiotic potential in Azov-Chernomoskaya royal fish by eliminating pathogens and increasing survival rate of fish [11] . Using the RAST server [49] , the protein-en- coding genes that were responsible for bacitracin-like antibiotics biosynthesis were identified [50] .
B. subtilis KATMIRA1933 was isolated from a dairy product [9] and it has been extensively consumed by people without any negative reported side effects. B. subtilis KATMIRA1933 was detected as an antimicrobial- producer strain. The bacteriocin subtilosin, which was a secondary metabolite of B. subtilis KATMIRA1933, showed selective antimicrobial activity against pathogens such as Gardnerella vaginalis but not against protective (beneficial) lactobacilli [51] [52] .
The antimicrobial activity of CFSs of B. subtilis KATMIRA1933 and B. amyloliquefaciens B-1895 against oral pathogens was reported in this study. These results attracted our attention moving toward evaluating the probiotic potentials of these bacilli. Safety tests were performed to ensure the innocuous side effects of the utilization of such microorganisms in food and medical applications.
An active area of oral health research has been aimed at the identification of natural products to either kill or attenuate pathogenicity of the key bacterial strains that contribute to oral infections such as caries and periodontal disease. Much of the effort has focused on finding plant-based compounds that are able to act as antimicrobials or bacteria-modulating compounds. An alternative active area of research is the identification of bacterial factors that can act as therapeutics, and as presented here, products of soil isolates that display antimicrobial properties against two classes of sub-gingival plaque organisms that have been associated with periodontitis and second-site infections such as brain or liver abscess, as in the case of S. intermedius.
As demonstrated here, B. subtilis KATMIRA1933 extract has anti-microbial effects against all P. gingivalis strains tests; these represent a combination of fimbrinated, afimbrinated, encapsulated, and non-encapsulated strains. Previous studies addressing P. gingivalis susceptibility to Bacillus extracts identified that only those without a capsule were susceptible, however, the researchers used different growth conditions which were presented here [53] . In our experience, P. gingivalis growth in liquid culture is suboptimal and there is mixed success in diluting into liquid media with insufficient CFU・mL−1 concentration. The measure is growth over starting inoculum, but we are not told if there was a control well with no subtilosin addition, which would have been the appropriate comparison [53] . Presented here regardless of capsule formation, we observed inhibition of P. gingivalis growth; however, we did observe that those strains without or with little capsule, 381 and 33,277 respectively, were slightly more susceptible to be killed by the extract, which was in agreement with the previous finding [53] .
Interestingly, Lys-gingipains of P. gingivalis do not inhibit activity of subtilisin as reported by Shelburne et al. [53] . However, the studied P. gingivalis strain 33,277 was already remarkably sensitive to subtilosin. It will be interesting to elucidate susceptibility of P. gingivalis W83 to subtilosin since this strain is lacking some proteases and is likely to be more sensitive to the antimicrobial protein. The presence of the protease on the surface is likely to degrade subtilosin. However, it was shown that the mutant lacking the proteolytic enzyme coding gene displayed similar MIC to the parent strain. Arg-gingipain absence failed to induce further sensitivity to subtilosin. The authors state that this could be due to the lack of arginine residues in proteins. Subtilosin contains lysine residues, however, it is not noted if the correct gingipain cleavage site is present in subtilosin, therefore the lys- gingpain also has the potential to be ineffective at cleavage. It should be noted that P. gingivalis has a host of proteases, and given the robust inhibition of growth, it would appear that the agent responsible for activity is not susceptible to proteolysis by P. gingivalis proteases. To confirm, this hypothesis would need to be formally tested.
Previous studies by Tsubura et al. [54] have demonstrated that Extraction 300E (E-300, AHC Co., Gunma, Japan), a preparation from the culture medium supernatant of Japanese soil B. subtilis isolate and the commercially available VITALREX (AHC Co., Gunma, Japan), a stable oral tablet of lyophilized B. subtilis DB9011, were both effective in reducing periodontitis levels in patients in comparison to controls [54] [55] . The BANA tests that were used to assay for periodontitis showed reduced levels in patients treated with the compounds; this test indicates that there are organisms that are able to hydrolyze the synthetic peptide benzoyl-DL-arginine- naphthylamide, which are attributed to the Gram negative anaerobes present in the subgingival plaque, of which P. gingivalis is a member. A reduction in BANA levels also coincided with a reduction in a number of target bacteria, including P. gingivalis and P. intermedia [55] .
It is being debated which tests may serve as a “minimum requirement” to characterize a putative probiotic’s safety and value [1] [56] [57] . Most agree that the antibiotic susceptibility of bacterial cells is the test of priority to identify if they are tolerant or sensitive to commonly prescribed antibiotics. Resistance of probiotics to antibiotics has both positive and negative impacts on human health. When bacterial resistance is intrinsic, it helps and supports the restoration of intestinal microbiota after a course of antibiotics that are administrated to the host for infection treatment [57] . At the same time, it is problematic when antibiotic resistance-genes in probiotics are transferable to other microbiota or pathogens leading to the appearance of new resistant strains [58] . In our study, we evaluated the antibiotic susceptibility, using a disc diffusion test, of the two Bacillus strains to nine antibiotics. The tested bacilli were more tolerant to bacitracin and streptomycin but susceptible to penicillin, ampicillin, erythromycin and chloramphenicol more than other used antibiotics. Also, tolerance of B. amyloliquefaciens B-1895 to tetracycline and oxytetracycline was higher than that of B. subtilis KATMIRA1933.
Bacitracin production by the Bacillus themselves reflects the natural resistance of Bacillus strains to these antibiotics. Bacillus resistance to bacitracin occurs either through the specific transporter protein, BcrABC, which takes bacitracin out of the cell [59] or by an undecaprenol kinase, which provides C55-isoprenyl phosphate, the BacA [60] . A putative bacitracin transport permease has been identified in Bacillus subtilis. This protein encoded by the B. subtilis bcrC (ywoA) gene is associated with bacitracin resistance [61] . Our data were in agreement with Senesi et al. [62] and Adimpong et al. [63] who found that all tested Bacillus strains were resistant to streptomycin and tetracycline at certain concentrations. Resistance of B. subtilis KATMIRA1933 to several antibiotics was not a surprising finding and it was confirmed by genome annotation data that was performed by Karlyshev et al. [64] for the studied bacilli using RAST analysis [49] . In addition to multidrug resistance efflux pumps encoding genes in B. subtilis KATMIRA1933, genes coding for resistance to vancomycin, fluoroquinolones, fosfomycin, and β-lactam antibiotics were also identified. Importantly, these studies referred to the fact that the probiotic bacilli that are used in animal and human food industries have shown multi-drug resistance behavior, especially toward streptomycin and tetracycline [65] - [67] . In the case of B. amyloliquefaciens B-1895, the β-lactamase gene, the streptothricin acetyltransferase-biosynthesis gene, and genes for resistance to fluoroquinolone and tetracycline antibiotics were also detected [50] .
Antibiotic resistance of bacterial species is either intrinsic or acquired by the transfer of a gene from plasmids, transposons, or the mutation of the bacterial gene [68] [69] . Mazza et al. [70] tested the resistance stability of antibiotic resistance markers existing in B. subtilis O/C, T, N/R, and SIN strains. Four therapeutic antibiotics were considered (chloramphenicol, tetracycline, rifampicin and streptomycin). They noticed that the resistance stability to tetracycline, rifampicin, and streptomycin existed for at least 200 generations without selective pressure. In vivo and in vitro studies explained the “absence of homologous transfer of resistance markers among the resistant strains” [70] . Bacillus species such as B. subtilis have been included on the Qualified Presumption of Safety (QPS) microbes list. The QPS list was generated including microbial taxonomic units in which the acquired antibiotic resistance-genes are absent [3] [71] .
In addition to antibiotic resistance, toxigenic potential such as the hemolytic activity [72] of Bacillus was evaluated in this study. It is known that hemolysin enzyme production is one of the virulence factors of pathogenic microorganisms. In whole blood agar, a weak hemolytic zone around B. subtilis KATMIRA1933, but not B. amyloliquefaciens B-1895, was observed and compared with the clearer zone of hemolysis produced by S. aureus, a positive control. No hemolytic activity of both B. subtilisKATMIRA1933 and B. amyloliquefaciens B-1895 was detected on defibrinated blood agar. Therefore, we assumed that the activity of B. subtilis KATMIRA1933 on the whole blood agar was fibrinolytic, not hemolytic action. To confirm our assumption, a fibrinolytic assay was performed for both tested bacilli. The clear zone of fibrinolytic activity produced by B. subtilis KATMIRA1933 was wider than the zone generated by the Bacillus strain B. amyloliquefaciens B-1895. The question that remains is why B. amyloliquefaciens B-1895, which has fibrinolytic activity like B. subtilis KATMIRA1933, did not produce the zone of hemolysis on the whole blood agar. However, hemolytic activity was determined in commercial human Bacillus species commonly used as probiotics [72] [73] . Hemolytic activity is a highly recommended test by EFSA to ensure that the bacterial strain was free of the toxigenic potential [74] . Although studies on fish and pigs reported that the hemolytic activity of microbial enzymes in vitro does not necessarily produce any negative effect in vivo [75] [76] , hemolytically active bacteria are not recommended as feed additives according to EFSA guidelines [71] . Based on our results, we propose the evaluation of bacterial hemolytic activity accompanied by the assessment of fibrinolytic activity.
A bacterial reverse mutation (Ames) assay was performed to evaluate the mutagenic potential and ensure safe utilization of Bacillus’ metabolites. The mutant frequency value confirmed that CFSs of B. subtilis KATMIRA1933 and B. amyloliquefaciens B-1895 were free from mutagenic substances. In this work, we used S. typhimurium K-6 TA1535 as his-mutant strain without using a metabolic activator (S9 mix). Mortelmans and Zeiger [30] , Vijayan et al. [77] , and Lupi et al. [78] found no significant differences in the number of revertant colonies of S. typhimurium TA1535 in the presence or absence of S9. The results of the Ames test were considered as primary data but did not guarantee that bacilli cell products do not have any mutagenic or carcinogenic active substances [79] . We agree that some supportive tests are needed to strengthen our finding, such as the Micronucleus assay, in vitro chromosomal aberration assay, and oral toxicity studies in rats to identify genotoxicity and clastogenicity [79] .
According to the WHO definition that probiotics produce health benefits in the host, proteolytic activity of the tested bacilli was evaluated and identified. Our data showed that B. subtilis KATMIRA1933 and B. amyloliquefaciens B-1895 behaved like many previously studied Bacillus species, showing a clear zone of proteolytic activity when they were inoculated on milk agar medium. Sevinc and Demirkan [80] found that utilizing skim milk agar for the qualitative test of protease is better than casein agar. The advantages of proteolytic enzymes have been reported in many studies [27] - [29] . In these studies, the importance of proteolytic enzymes includes the activation of regeneration processes, the enhancement of fibrinolytic activity in the plasma, the enhancement of normal digestion processes, and degradation of allergic and chemical compounds.
The ability of probiotics to co-aggregate and auto-aggregate is considered an advantageous characteristic feature. Their adhesion to a pathogenic organism can facilitate the elimination of the organism from the body and its ability to self-aggregate gives it an advantage in a competitive environment. The microorganisms employed in this study were chosen as common pathogenic organisms found in products of consumption. Two mathematical analyses of the coaggregation data were evaluated, Method 1 was described by Ledder et al. [35] and Method 2 was described by Handley et al. [45] . The more comprehensive method for mathematical interpretation of optical density data was chosen on the basis of two requirements. Foremost, the calculated percentage of coaggregation had to make sense in its application. A mathematical form of analysis may often fall short when the parameters of a biological system must be taken into account, not quite “fitting” in a representative attempt. Second of all, the percentages that better reflected the visual analysis were identified. The extent of coaggregation and auto-aggregation was determined from the data that was found using the more appropriate method.
Method 1 appeared to be most appropriate for the data analysis in this study, for it agreed with the microscopic (visual) observations (Figure 1). Method 2 failed to adequately reflect the adhesion in a number of mixtures of microorganisms that were easily observed microscopically. For instance, some Method 2 derived values were <0, although coaggregation was clearly visible under the microscope (Figure 1). This method error could be due to any number of factors that affected the microorganisms, if the equation reflects a specific state. Method 1, however, gave percentages of coaggregation that more closely resembled the scores given during visual analysis, signifying that it was more appropriate for the data at hand.
After 2 h, B. amyloliquefaciens B-1895 adhered most to P. aeruginosa and E. coli more than other bacterial cells. Though these data did ideally match the coaggregation percentages of B. amyloliquefaciens B-1895 with P. aeruginosa and E. coli, which were found to have 31.5% and 31.4%, respectively, at this optical density when method 1 was applied (Table 6).
B. subtilis KATMIRA1933 adhered mostly to E. coli and P. aeruginosa, but less to S. enterica, S. aureus, and L. monocytogenes (Table 8). Similarly, B. subtilis KATMIRA1933 mostly coaggregated with the above-menti- oned bacterial cells. Both Bacillus strains were found to have auto-aggregating abilities after 2 h, especially B. amyloliquefaciens B-1895 (86%), which was also reflected in the visual analysis (data not shown).
According to the percentages of auto-aggregation (Table 7), B. amyloliquefaciens B-1895 showed greater instances of auto-aggregation than B. subtilis KATMIRA1933, at a more significant level, during 8 h. For B. amyloliquefaciens B-1895, the highest percentage of coaggregation was noticed with E. coliand P. aeruginosa and the lowest was with S. aureus (Table 8). B. subtilis KATMIRA1933 was highly coaggregated with E. coli, P. aeruginosa, and L. monocytogenes but less with S. mutans (Table 8).
Spores of B. subtilis KATMIRA1933 and B. amyloliquefaciens B-1895 showed high tolerance to different pH values of 2.0, 2.5, and 3.0 that simulate gastric conditions. The survival rates (%) of SFU in various acidic conditions during 4 h of incubation were either similar or less than 1-log reduction of the viable spore count when compared with a control (at time zero), and more than 80% of Bacillus spores survived in 0.3% bile salts (Data not shown). Duc et al. [81] stated that not all the spores of Bacillus probiotic strains were tolerant to gastric acidity and bile compounds in the upper gastrointestinal tract. Diversity in acid and bile tolerance of Bacillus species spores has been detected. When B. coagulans Unique IS-2 spores were evaluated by Sudha et al. [82] , a 2-log reduction of SFU・mL−1 was identified after exposure to pH 1.5 and a 1-log reduction at both pH 2.0 and 3.0 at 3 h. Also, three strains of B. coagulans (BCI4 LMAB, CIP5264 and CIP6625) were tested by Hyronimus et al. [47] , who reported the high susceptibility of these strains to acid (pH 2.5) and 0.3% bile after 3 h incubation. Our findings were in agreement with Guo et al. [83] who found that the number of B. subtilis MA139 that exerted probiotic potential in the gastrointestinal tract of pigs, was steady (unaffected) at pH 2.0 and in nutrient broth supplemented with 0.3% bile for 3 h. The high tolerance of Bacillus spores to gastric acidity and bile salts of the proximal intestine, compared with vegetative cells, are assorted properties required for the selection of probiotics [84] and are promising in acidified food packaging and oral pharmaceutical applications.
Germination of Bacillus spores in the intestinal environment has been mentioned in many studies [73] [85] [86] . Studies such as Barbosa et al. [87] , Duc et al. [81] and Fakhry et al. [88] were conducted to evaluate the tolerance of vegetative cells of Bacillus to acids and bile salts. Our data were in agreement with these studies in which high susceptibility of cells, in contrast to spores, to both conditions was reported. Studies of spore structures are required to determine the correlation with the susceptibility of some Bacillus spores to acids or bile salts. Evaluation of the survivability of spores in the presence of gastrointestinal tract enzymes, lysozymes, acids and bile salts [84] [86] need to be conducted in vivo to reflect the real assessment of Bacillus stability (sporulation or germination) under such conditions.
Before the establishment of probiotic capacity, other properties of B. subtilis KATMIRA1933 and B. amyloliquefaciens B-1895 should be further evaluated such as cell adhesion, hydrophobicity, and genotoxicity. To conclude, safety of the studied bacilli to human health should be established and completely confirmed due to the exciting potential of Bacillus for personal care, food and medical applications.
Acknowledgements
M. L. C. is grateful to Jiangsu Sinoyoung Biopharmaceutical Co., Ltd for their continuous support. V. A. C., M. S. M. and A. B. B. were supported by the Ministry of Education and Science of Russian Federation, project No. 6.1202.2014/K.
Conflict of Interest
The authors declare no conflict of interest.
NOTES
![]()
*Corresponding author.