Received 3 March 2016; accepted 26 April 2016; published 29 April 2016

1. Introduction
Alfalfa (Medicago sativa L.) is important legume forage that contains a variety of vitamins and high protein content and thus is a key component of the animal husbandry all over the world. Lucerne is a good plant model for studies of genetics. Many reports have been documented regarding genetic diversity of Medicago sativa using different traits: morphological and yield [1] - [3] , histological [4] and allozyme markers [5] .
Neutral, DNA-based molecular markers allow a more precise and environment-independent way to evaluate the genetic diversity of a particular species. Several studies have been done on alfalfa using DNA-based markers like AFLP [6] [7] , RAPD [8] - [13] , RFLP [14] - [17] and SSR [18] - [20] . These studies have shown that alfalfa was characterized by a high inter-population variability.
Among different markers (morphological, cytological biochemical and DNA), markers based on DNA are independent from environmental effects, unlimited in number and show high level of polymorphism.
Random Amplified Polymorphic DNA (RAPD) is generally favored because of its sensitivity, coupled with the fact that DNA sequence information is not required for primer design, no radioisotope labeling is needed for sample detection, and only a small amount of template DNA is required [21] [22] . RAPD is a molecular marker widely used in genetic variability analyses due to its simplicity and speed, dispensing previous genetic information of the sample under study [23] ; although no locus specific information is presented [24] . Basically, it’s a variation of the PCR amplification technique, using a simple primer of arbitrary sequence that will amplify a random region of the analyzed genome. Even though the reproducibility of this marker has been previously questioned [25] , adjustment in the technique has made the obtainment of reproducible results possible [26] permitting the use of this markers in many types of genetic analysis.
ISSR markers are generated via PCR reactions with a single primer designed from repetitions of two or three nucleotides anchored in a sequence of one to three nucleotides that aim to eliminate slippage related artifacts [27] . The amplified regions represent the sequence between two microsatellite sites and the absence of bands is interpreted as a divergence of the primer or loss of a locus by the deletion of the SSR sites or chromosomal rearrangement. Some advantages related to this marker are the small quantity of DNA required for the reactions, the small number of PCR reactions, the hyper variability of the bands patterns and their easy detection, besides the high PCR reactions annealing temperatures that reduce the quantity of artifacts and errors [28] .
The objective of this study was to investigate the differentiation level among a set of alfalfa populations with RAPD and ISSR markers, and to examine the relationship between these populations collected from tow semi arid regions (the South of Tunisia and the North West of China) and their genomic diversity in order to complete genetic resources conservation and the evaluation of native germplasm of lucerne and constitute a base for breeding program.
2. Materials and Methods
2.1. Plant Material
Twenty alfalfa (Medicago sativa L.) populations were involved in this study, including nineteen from South Tunisia and one from North Ouest of China. The description of the 20 alfalfa genotypes used in this study was resumed in Table 1.
2.2. Genetic Diversity Analysis
2.2.1. DNA Extraction
Young leaflets were harvested on each plant and total genomic DNA was purified from frozen young leaves according to the procedure described by [29] with minor modifications. The genomic DNA concentration was estimated spectrophotometrically and its integrity was checked by analytical (2%) agarose minigel electrophoresis [30] .
2.2.2. RAPD Assay
Five random decamer primers were used for DNA amplifications (Table 2). Polymerase chain reaction for each sample was performed in a total volume of 20 µl. The PCR (Polymerisation Chain Reaction) reaction mixture contained 50 ng genomic DNA, 1.5 mM MgCl2, 0.2 mM dNTPs, 0.5 µM primer and 1U Taq polymerase (Promega), 10× Taq polymerase buffer. PCR amplifications were performed in a Thermocycleur TC32/80 Cleaver scientific LTD system. The PCR conditions included initial denaturation at 94˚C for 5 min, followed by 45 cycles: denaturation at 94˚C for 1 min, annealing at Tm˚C of each primer (1 min), extension at 72˚C for 1 min
![]()
Table 1. Tunisian and Chinese alfalfa populations studied and their geographical origin.
![]()
Table 2. List of the selected RAPD primers and the polymorphisms obtained on 20 alfalfa populations.
with final extension at 72˚C for 7 min. Amplification products were loaded on 1.8% agarose gel, by electrophoresis in TBE buffer, stained with ethidium bromide, visualized by illumination with ultraviolet light and photographed under UV light, by a Bioprint (Kaiser RS1, Germany).
2.2.3. ISSR Assay
Five primers were used for PCR amplification (Table 3). PCR was carried out at 20 μL final volume using 25 ng of genomic DNA containing 4 μL of 5 × Green GoTaqR (pH 8.5, 7.5 mM MgCl2), 100 μM dNTPs, 150 pmol random primer, and 1.2 units of Taq DNA polymerase. The mixture was brought up to 20 μL by adding
![]()
Table 3. List of the selected ISSR primers and the polymorphisms obtained on 20 alfalfa populations.
sterilized distilled water. The mixture was amplified in a thermal cycler (GeneAmpR PCR System 9700) that was programmed for 1 cycle of initial denaturation at 94˚C for 5 min; 35 cycles of 94˚C for 1 min, followed by specific annealing temperature for 55 s, and ending with an extension at 72˚C for 1 min; and a final extension cycle at 72˚C for 7 min. The PCR machine was adjusted to hold the product at 4˚C. The PCR products and 1 kb DNA ladder were electrophoresed on 2% agarose gel (stained with EtBr). The separated fragments were visualized with an ultraviolet (UV) transilluminator.
2.3. Data Analysis
Each gel after electrophoresis and staining was analysed by scoring manually the present (1) and absent (0) polymorphic RAPD and ISSR bands in individual lines for each primer and the values were used to compile binary data matrix.
For all primers combination, the total number of bands was determined and only the polymorphic ones were taken into account in this study to estimate the percentage of polymorphic bands (%PB).
The similarity matrix was calculated with Jaccard’s coefficient [31] . The matrix was then analyzed with the Neighbor-joining program using PHYLIP software (Phylogeny Inference Package, version 3.5c) [32] to construct a dendrogram between 19 Tunisian and 1 Chinese alfalfa populations using the unweighted pair group method with arithmetic averaging (UPGMA) algorithm.
The ability of the most informative primers to discriminate among populations was assessed by calculating the resolving power (Rp) [33] which has been reported to correlate between accessions. Evaluation of the Rp was performed according to the formula of [34] : Rp = ΣIb, where: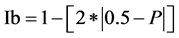 , Ib is the informativness band and P is the proportion of the accessions containing the I band. Average Informativeness band (AvIb) as a measure of closeness of a band to be present in 50% of the genotypes in this study, was also calculated.
, Ib is the informativness band and P is the proportion of the accessions containing the I band. Average Informativeness band (AvIb) as a measure of closeness of a band to be present in 50% of the genotypes in this study, was also calculated.
On the other hand, a correspondence factorial analysis (CFA) was performed by computing the binary matrix with XLSTAT version 14.
3. Results
3.1. RAPD Markers in Determining Genetic Variation of Alfalfa Populations
In order to detect genetic variation between 20 alfalfa populations, PCR amplifications were performed with RAPD primers. A total of five primers were screened in this study and produced discrete and reproducible amplified DNA fragments (Figure 1). The five RAPD primers resulted in 44 different amplification products with 22 polymorphic RAPD bands for the 20 populations (Table 2).
Different levels of polymorphisms were detected since as the percentage of polymorphic bands (%PB) ranged from 14.28 for W07 to 77.77 for AT primers, with an average of 46.02%.
The percentage of polymorphic bands detected by RAPD marker was 50% and the average of polymorphic bands per RAPD primers was 10%. The largest number of polymorphic bands (8) was produced with AF14 primer, and the least number of polymorphic bands (1) was obtained by W07. The 5 polymorphic primers exhibited variation with regard to Average Informativeness band (AvIb) and resolving power (Rp). However, estimates of the resolving power (Rp) showed a high rate of collective Rp (10.1), with an average of 2.02.
![]()
Figure 1. RAPD bands amplified with five arbitrary primers: (A) AF 14, (B) C1, (C) AX16, (D) AT and (E) W07, using DNA of 20 Tunisian and Chinese alfalfa samples. Numbers represent the populations according to Table 1. MM: Molecular Marker.
Moreover, as reported in Table 2, the Average Informativeness band (AvIb) values varied from 0.2 showed by the primer C1 to 0.9 showed by the primer W07, with an average of 0.522.
The similarity matrix (Table 4) showed that the highest similarity (0.87) was for the populations P4, P5, P10, P13 and P18, P1, P14, P15 and P16, P2 and P11. The lowest similarity (0.55) was between the populations P21, p20, P4, P1, P11, P18, P10 and P13.
The derived UPGMA dendrogram illustrated in Figure 2 has clustered the 20 populations into seven main groups. The three first groups, labeled (I, II, III), successively, consisted of “Xinjiang Daye” (Provenance Chine), P20: Stiftimia (Provenance Kébili) and P2: Chenchou (Provenance Gabès) et P11: Bouhlel (Provenance Tozeur). The fourth group labeled (IV) consisted of P1: Kattana (Provenance Gabès), P14: Zaafarane, P15: Nouael et P16: Jersine (Provenance Kébili) populations. The fifth group (V) consisted of P4: Chenenni1, P5:
![]()
Table 4. Similarity matrix among 20 alfalfa populations based on data from 5 RAPD primers (44RAPD markers).
Chenenni2, P6: Chenenni3 (Provenance Gabès), P10: Essdada, P13: Hamma jerid (Provenance Tozeur) et P18: Limaguess (Provenance Kébili) populations. The sixth (VI) contained P3: Tboulbou, P7: Metwia, P8: Ghannouch et P9: Zerkine (Provenance Gabès) populations. Finally the last group (VII) consisted of tow populations: P12: Dgach (Provenance Tozeur) et P17: Elgolaa (Provenance Kébili).
To evaluate the information contained in experimental data, correspondence factorial analysis (CFA) was applied considering all the monomorphics and polymorphics bands obtained simultaneously by the 5 RAPD primers. The first 2 axis accounted for 29.06%. Axis 1 explained 15.35% of the inertia, however the axis 2 explained 13.71% of the inertia. The dispersion of alfalfa populations on the plan defined by axes 1 and 2 showed seven clusters of populations (I, II, III, IV, V, VI and VII). Theses clusters were similar to these obtained by the dendrogram illustrated in Figure 3 and contained the same populations. Grouping of lucerne genotypes using CFA confirmed the result obtained using UPGMA method based on RAPD data. Tunisian and Chinese alfalfa populations were clustered independently of both geographic origins.
3.2. Genetic Diversity and Genotypes Relationships Obtained by ISSR Markers
In the second part of our experiment, the genetic variation between 20 alfalfa populations was detected with five ISSR primers. These five primers were screened in this study and produced discrete and reproducible amplified DNA fragments (Figure 4). The five ISSR primers resulted in 51 different amplification products with 33 polymorphic RAPD bands for the 20 populations (Table 3).
Different levels of polymorphisms were detected since as the percentage of polymorphic bands (%PB) ranged from 20 for A12 to 95.56 for UBC 890 primers, with an average of 57.78%.
![]()
Figure 2. Dendrogram of the 20 Tunisian and Chinese alfalfa populations constructed by UPGMA methods using the similarity matrix generated by the Jaccard coefficient based on RAPD marker.
![]()
Figure 3. Plot of 20 Tunisian and Chinese alfalfa populations according to the tow first axes of the correspondence factorial analysis (CFA) based on 5 RAPD primers.
![]()
Figure 4. RAPD bands amplified with five primers: (A) A1, (B) Am2, (C) UBC 890, (D) A12 and (E) UBC 818, using DNA of 20 Tunisian and Chinese alfalfa samples. Numbers represent the populations according to Table 1. MM: Molecular Marker.
The percentage of polymorphic bands detected by RAPD marker was 64.7% and the average of polymorphic bands per RAPD primers was 12.94%. The largest number of polymorphic bands (22) was produced with UBC 890 primer, and the least number of polymorphic bands (1) was obtained by A12. The 5 polymorphic primers exhibited variation with regard to Average Informativeness band (AvIb) and resolving power (Rp). However, estimates of the resolving power (Rp) showed a high rate of collective Rp (11.4) with an average of 2.28.
Moreover, as reported in Table 3, the Average Informativness band (AvIb) values varied from 0.29 showed by the primer UBC890 to 0.7 showed by the primer A12, with an average of 0.624.
The similarity matrix (Table 5) showed that the highest similarity (0.97) was for the populations P4, P5, P6, P10, P13 and P18; P1, P14, P15 and P16, P2 and P11. The lowest similarity (0.6) was between the populations P21, P4, P1, P14, P18, P20 and P9.
![]()
Table 5. Similarity matrix among 20 alfalfa populations based on data from 5 ISSR primers (51ISSR markers).
The derived UPGMA dendrogram illustrated in Figure 5 has clustered the 20 populations into five main groups. The three first groups, labeled (I, II), successively, consisted of P21: Xinjiang Daye (Provence Chine) and P20: Stiftimia (Provence Kébili). The third group labeled (III) consisted of P2: Chenchou, P3: Tboulbou, P6: Chenenni 3, P8: Ghannouch and P9: Zerkine (Provence Gabès), P17: Elgolaa (Provence Kébili). The fourth group labeled (IV) consisted of P1: Kattana, P4: Chenenni 1, P5: Chenenni 2, P7: Metwia (Provence Gabès), P10: Essdada, P12: Dgach and P13: Hamma Jerid (Provenance Tozeur) and P18: Limaguess (Provenance Kébili). Finally the last group (V) consisted of tow populations: P15: Nouael and P16: Jersine (Provence Kébili).
To evaluate the information contained in this experimental data, correspondence factorial analysis (CFA) was applied considering all the monomorphics and polymorphics bands obtained simultaneously by the 5 ISSR primers (Figure 6). The first 2 axis accounted for 45.07%. Axis 1 explained 24.60% of the inertia, however the axis 2 explained 20.47% of the inertia. The dispersion of alfalfa populations on the plan defined by axes 1 and 2 showed five clusters of populations (I, II, III, IV and V). Theses clusters were similar to these obtained by the dendrogram illustrated in Figure 4 and contained the same populations. Grouping of lucerne genotypes using CFA confirmed the result obtained using UPGMA method based on ISSR data. Tunisian and Chinese alfalfa populations were clustered independently of both geographic origins.
4. Discussion
A study is presently carried out on the genetic variability available in this specie. Codominant Molecular markers such as RAPD and ISSR were used to investigate the genetic diversity and to study the relationships between
![]()
Figure 5. Dendrogram of the 20 Tunisian and Chinese alfalfa populations constructed by UPGMA methods using the similarity matrix generated by the Jaccard coefficient based on ISSR marker.
![]()
Figure 6. Plot of 20 Tunisian and Chinese alfalfa populations according to the tow first axes of the correspondence factorial analysis (CFA) based on 5 ISSR primers.
20 populations of Tunisian and Chinese alfalfa. However in both of RAPD and ISSR markers, a set of 20 populations were analyzed in this study (Table 1). A total of 10 primers were screened for RAPD and ISSR PCR analysis. They were useful for characterizing the samples and produced strongly amplified polymorphic bands. The selected primers generated an appropriate amplification pattern with clear, consistent, and reproducible bands. In the present study the total number of amplified bands for RAPD markers was 44 of which 22 were polymorphic. Moreover, the average polymorphism displayed by this marker was 50%. Many reports have been documented regarding genetic diversity of Medicago sativa L. using RAPD markers [8] - [13] .
However, the total number of amplified bands for ISSR markers was 51 of which 32 were polymorphic. Moreover, the average polymorphism displayed by this marker was 64.7% however, nothing is know about the genetic diversity available within or between alfalfa populations based on ISSR marker except the previous study concerning the genetic diversity of some Mediterranean alfalfa populations (Medicago sativa L.) using ISSR marker [35] .
The total variability that may be due, mainly, to interaction of several factors; 1) The reproductive aspect of the alfalfa (outcrossing); 2) the uncontrolled introduction of the varieties improved in Tunisia [36] ; and 3) the insufficiency of the seeds [2] .
The average polymorphism displayed by ISSR (64.7%) was higher than that by RAPD (50%), which suggested that the ISSR markers were superior to RAPD markers in the capacity of revealing more informative bands in a single amplification. ISSR markers proved, however, to be more useful, due to the high polymorphic percentage between alfalfa populations. ISSR results are more realistic comparing to RAPD results. Researchers who have compared RAPD and ISSR methods have found that ISSR markers exhibit higher levels of polymorphism or reproducibility compared with RAPD markers [37] - [40] .
The dendrogram obtained by the Unweighted Pair Grouped of aggregation (Method Average (UPGMA) based on Jaccard distance and the Correspondance Factorial Analysis revealed a considerable variability between populations. However these analyses based on RAPD or ISSR markers showed respectively seven and five clusters of alfalfa populations. In the majority of group, the populations were different provinces. Genetic diversity observed in our analysis is not structured according to the geographic origins of population because the groups obtained in a classification aren’t related to provinces. Benabderrahim et al. [2] show that the distribution of twenty cultivated populations of lucerne (Medicago sativa L) collected from different oasis of Tunisian south and evaluated for morphology and yield is not according to their geographic origins.
5. Conclusion
The results obtained here showed the possibility of genetic diversity analysis of alfalfa populations, based on dominant RAPD and ISSR markers treated with UPGMA method. RAPD and ISSR tools are very important for explaining genetic diversity and alfalfa population structures. The observed differences between studied populations should be studied further using other molecular approaches such as: RFPL, AFLP and SSR, these markers could be used in breeding program for improving vegetative and reproductive traits and to detect QTL (quantitative traits locus) for physiological traits related to abiotic stress adaptation.
NOTES

*Corresponding author.