Association between UCP Gene Polymorphisms and Growth, Breeding Value of Growth and Reproductive Traits in Mazandaran Indigenous Chicken ()
Received 20 October 2015; accepted 9 January 2016; published 12 January 2016

1. Introduction
Low energy expenditure, in addition to increased energy intake, has been a major cause of future weight gain, and variations in energy expenditure may be one of the underlying sources of variation in body weight [1] . Various determinants, including body composition, hormonal levels, activity of the sympathetic nervous system, and genetics are responsible for differences in metabolic rate among individuals [2] . A mitochondrial protein called uncoupling protein plays an important role in generating heat and burning calories by creating a pathway that allows dissipation of the proton electrochemical gradient across the inner mitochondrial membrane in brown adipose tissue, without coupling to any other energy-consuming process. This pathway has been implicated in the regulation of body temperature, body composition, and glucose metabolism [3] . The uncoupling protein (UCP) gene is one of the many genes that maybe related with growth traits, breeding value of growth and reproductive traits. UCP3 is the member of uncoupling proteins (UCPs) family of mitochondrial transporters, all of which are known to uncouple oxidative phosphorylation via proton leakage from the inner mitochondrial membrane. Thermoregulation and energy metabolism are potentially regulated by these mitochondrial proteins [4] . The uncoupling protein gene (UCP) is a candidate gene for growth traits, breeding value of growth and reproductive traits because it may contribute to the proton leak that occurs in mitochondria and partially uncouples the oxidative phosphorylation, thus releasing chemical energy as heat [5] . Uncoupling protein (UCP) gene family involved in energy metabolism of body, and had significant effects on energy balance related traits, such as body weight resting metabolic rate, and efficiency of food conversion etc. [6] . Liu et al. [7] found UCP-3 gene polymorphisms may contribute to body mass index (BMI) in the Caucasian population. Rudofsky et al. [8] found that human UCP-3 gene involved in energy balance and was associated with obesity and diabetes. UCP2 gene is expressed extensively in various tissues, like white adipose tissue, pancreas, skeletal muscle, and liver, in human [9] . Both environmental and genetic factors may have contribution toward susceptibility of growth traits, breeding value of growth and reproductive traits. Genetic diversity in indigenous breeds is a major concern considering the necessity of preserving what may be a precious and irreplaceable richness, regarding new productive demands. Conservation should be based on a deep knowledge of the genetic resources of the specific breed. Therefore, it is important to try to characterize genetically indigenous breeds [10] . A species without enough genetic diversity is thought to be unable to cope with changing environments or evolving competitors and parasites and also the ability of a population to respond adaptively to environmental changes depends on the level of genetic variability or diversity it contains. Therefore, studies of population genetic diversity and the structure of population within and between species may not only illustrate the evolutionary process and mechanism but also provide information useful for biological conservation of the Bovidae family, sheep, goats, birds and so on [11] . Molecular markers are increasingly used for the study of genetic diversity of populations in recent years [12] . Therefore, in the present study, we aimed to investigate association of polymorphism of UCP2 gene with growth traits, breeding value of growth and reproductive traits in subjects from Iran.
2. Material and Methods
2.1. Experimental Population and Sampling
Breeding station of Mazandaran Indigenous Chicken is located at 28 km far from Sari, the provincial capital of Mazandaran state, located in the north of
Iran
. In 1986 about 5000 cocks and hens were purchased from rural regions across the Mazandaran province and kept in a quarantine farm for a year. From those, about 2500 birds of two sexes were kept to produce hatching eggs and chicks produced from these eggs were transferred to the station in 1988. Since then birds have been individually tagged and trap nest has been used for pedigree recording. Parents of each generation (about 100 cocks and 800 hens) are selected among 6000 pedigreed and performance recorded birds produced each generation. In August 2009, a total of 205 blood samples from Mazandaran Indigenous Chicken, including 10 males and 195 females, were collected. Individuals were reared in Native chicken breeding station of Mazandaran and they belonged to generation 17 of the breeding station pedigreed animals. Individuals of this generation were developed by crossing 80 sires and 751 dams from generation16. Approximate 1 ml blood per chick from plumage veins was collected and kept in a tube containing anticoagulant EDTA (Ethylenediaminetetraacetic acid). All samples were delivered back to the laboratory in an ice box. The genomic DNA was extracted from white blood cells using standard salting out procedure described by Abadi et al. [13] . The DNA samples were dissolved in TE buffer which was made from 10 mMTris-Cl (pH 7.5) and 1 mM EDTA (pH 8.0) and were stored at 20˚C for use.
2.2. Primer Synthesis and PCR-RFLP Reactions
The primers were designed on the basis of DNA sequence of the UCP (Accession: AF433170) using the oligonucleotide design tool Primer 5.0 software (F: 5’-TACCCCAAGCATGCAGAACTCA-3’ and R: 5’- GGAACCGCACCTTGACCAC-3’). Both PCR reactions were performed in a 20 µl mixture containing 10 pmol primers, 200 lMdNTP (deoxyribonucleotide triphosphate), 2 µl 10 × reaction buffer which contained 1.5 mM MgCl2, 1 unit of Taq-DNA polymerase (Promega, Madison, WI), and 50 ng genomic DNA as template. PCR method was used to optimize the reaction accuracy: 94˚C for 5 min, 35 cycles of 94˚C for 30 s, annealing at 55˚C for 60 s, 72˚C for 60 s, and a final extension at 72˚C for 7 min. PCR products were electrophoretically separated on 2% agarose gel (5 V/cm), stained with ethidium bromide. PCR products were digested by 10 units of Hha1 restriction enzymes (Fermentase,
Lithuania
), 6 ml of PCR product, 1.4 ml of Tango buffer and 2 ml nuclease-free water. The final volume of 10 ml was incubated in 37˚C for 12h. The fragments were separated a 3.5% agarose gel stained with ethidium bromide.
2.3. Traits Measured
Whole information data file (18 successive generations) consisted of three fixed effects (generation, sex and hatch) and 11 recorded traits including body weight at hatch (BW1), body weight at age of 8 (BW8), 12 (BW12) weeks, body weight at sex maturation (WSM), age at first egg (ASM), egg number (EN), first egg weight EW1), average egg weight at age of 28 (EW28), 30 (EW30), 32 (EW32) weeks and average egg weight for the first 12 weeks of production (EW12). BW1, BW8 and BW12 have been measured in both male and female chicken. Also, three combined traits consisting of average of EW28, EW30 and EW32 (AV), intensity of egg production (EINT = (egg number/days recording) × 100) and egg mass (EM = EN × EW12) were calculated to use in the analyses (Table 1). During 18 generations, genetic evaluation of the birds for body weight at 8 weeks, age of the hens at first egg, average egg weight and total number of eggs laid during first 12 weeks after flocks maturity (when 5% of the flock are in egg production) have been performed. Economic indexes are calculated for these traits and birds of two sexes are selected based on their aggregate genotypes for these traits. The goals of the breeding station on the one hand are to increase body weight, egg weight and egg number and on the other hand, to decrease age at first egg.
2.4. Statistical Analyses
Pedigree and data file were prepared using Visual FoxPro 9.0 software, the relational data base management system. SAS9.1 package was used to carry out descriptive statistics and fitting model. Fixed effects and their interactions were considered in animal model that provided having significant effect. Genetic analyses were performed using ASREML software [14] . Breeding values of growth and egg production traits was estimated using the BLUP based on an animal model with a relationship matrix. The used models in matrix notation were as follows:
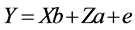
where, Y is the vector of observations; b the vector of fixed effects of generation, sex and hatch; a the vector of random direct genetic effects; e the vector of random residual effects; X and Z are incidence matrices relating the observations to the respective fixed and direct genetic effects. Calculation of gene frequency was based on direct gene count method by:  and standard error of frequency was calculated as (p(1 − p)/2n)1/2, where n is the sample size, p is the frequency of A or G allele. Marker-trait association analyses were conducted using following model in GLM procedure of SAS9.1 software. The significant differences of least squares means were tested with Tukey-Kramer’s multiple range tests, and a P-value of ≤0.05 was considered statistically significant.
and standard error of frequency was calculated as (p(1 − p)/2n)1/2, where n is the sample size, p is the frequency of A or G allele. Marker-trait association analyses were conducted using following model in GLM procedure of SAS9.1 software. The significant differences of least squares means were tested with Tukey-Kramer’s multiple range tests, and a P-value of ≤0.05 was considered statistically significant.
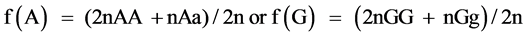
Yijk is the estimated breeding values of traits, µ the population mean, Mi the fix effect of genotypes, eijk the residual random error. There was no significant interaction between the genes additive effects.
3. Results
3.1. Analysis of the Polymorphisms in the UCP Gene
Five variations in the UCP gene are shown in Table 1 with data obtained from 190 chickens. A single nonsynonymous SNP was a C > T substitution at nt position 1270 in exon-3 replacing Ala > Val. A PCR-RFLP test using HhaI restriction enzyme was designed to screen the UCP Ala118Val polymorphism (Table 2). Figure 1 shows the electrophoretic gel pattern of the HhaI PCR-RFLP test where lanes 6 and 2 are Ala/Val heterozygotes with or without the 19-bp deletion, respectively. An additional restriction cut site was created within the PCR- amplified region by t > C silent substitution for cysteine at nt position 1316 in exon-3 that produced the second band from the top as seen in Figure 1 for lane 5. The digested allele as allele ‘‘T’’, and the undigested allele was
![]()
Figure 1. The electrophoretic gel patterns of the HhaI PCR- RFLP. M is Ladder, PCR is Fragment of PCR product, lanes 2 and 6 are heterozygote and other lanes are homozygote.
![]()
Table 1. Statistical description of data set for growth and egg production traits.
BW1, BW8, BW12 = Body weight at birth, 8, 12 weeks of age, WSM = Body weight at sexual maturity, ASM = Age at first egg, EN = Egg number, EW1 = Weight of first egg, EW28, EW30 and EW32 = Average egg weight at 28, 30 and 32 weeks of age respectively, EW12 = Average egg weight for first 12 weeks of production, AV = Average for EW28, 30 and 32, EM = Egg mass (=EN × EW12), EINT = Egg production intensity (= Egg number/days recording) × 100).
![]()
Table 2. Summary of variations in UCP gene.
identified as allele ‘‘C’’. Three genotypes TC (Ala/Val), TT (Ala/Ala) and CC (Val/Val), were detected after PCR- RFLP analysis of the UCP gene. The allele frequencies were 0.21 for T and 0.79 for C, and the genotypic frequencies were 0.18, 0.12, and 0.70 for TC, TT and CC, respectively.
3.2. Relationship between Polymorphisms and Growth, Breeding Value of Growth and Reproductive Traits
The association between the AF433170: c. C1270T polymorphisms in the UCP gene and growth, breeding value of growth and egg production traits are shown in Table 3 and Table 4, respectively. UCP gene, the Missense mutations were significantly linked to ASM (P < 0.05) and also highly significantly linked to breeding value of ASM (P < 0.05). In this population, ASM was higher in the heterozygote (UCP Ala/Val) compared with the homozygote (UCP Val/Val).
4. Discussion
The study of candidate genes is one of the primary methods to find specific genes related to economic traits in farm animals. UCP involved in regulation of energy metabolism has important physiological effects on economical important traits of farm animals. Gene polymorphisms can be used for improvement of the production traits by genetic selection, if the allelic association with the traits is determined. The chicken UCP gene was first discovered by Raimbault et al. [15] . Its function is still not clear, though there has been some research about it since its discovery. The nucleic acid sequence of avian UCP is highly homologous to both mammalian UCP2 and UCP3 [16] . The current hypothesis on the function of these two UCPs involves regulation of fatty acid oxidation, especially during metabolic stress [17] , possibly by transporting fatty acid ions out of the mitochondria [18] . Two significant associations (P < 0.05) were observed with growth, breeding value of growth and reproductive traits in this study (Table 3 and Table 4). Sherman et al. [19] observed significant association between a SNP (A/G) in bovine UCP-3 gene intron-3 and average daily gain and partial efficiency of growth in 9 Continental British hybrid beef steers. Han [20] reported the existence of three SNPs (G/A, C/T and G/T) in exon 3 and four genotypes in Chinese Qinchuan cattle. Association studies showed that the genotype AA had higher slaughter weight, carcass weight, carcass length, eye muscle area, water holding capacity, and marbling than other genotypes; the genotype AB had higher back fat than other genotypes. Li [21] found that the allele A of UCP-3/BglI polymorphism had higher frequencies than allele B in Nanyang, Luxi, and Yanbian cattle. Association analysis showed the superiority of the AA genotype over the AB genotype for the content of b-globin in Luxi cattle. Fasting, which is associated with a major reduction in total body energy expenditure, is associated with an increase in the expression of UCP2 and UCP3 [22] . However, it was also reported that UCP2 and UCP3 genes were unlikely to have a substantial effect on variation in obesity phenotypes, in particular US Caucasian human population [23] . In animal and human studies, UCP2 mRNA levels have been correlated with plasma-free fatty acid concentrations [24] . In adipose tissue of obesity-resistant mice, UCP2 expression is increased 2-fold when compared with obesity-prone animals [25] . Independent studies have identified quantitative trait loci for obesity or body fat in mice that coincide with the genomic location of the UCP2 gene [26] [27] . The SNP in the exon included synonymous and nonsynonymous polymorphisms. At any given position in a DNA sequence, a nucleotide can be substituted by any of the 4 nucleotide bases and may result in biallelic SNP. This occurs due to the low substitution rate of single nucleotides, estimated to be between 1 × 10−9 and 5 × 10−9 per nucleotide per year at neutral positions in mammals [28] . Based on these numbers, the probability of 2 independent base changes occurring at a single position is very low [28] . A large variation in UCP gene sequences in the studied population was found to reside in the first half of the gene. In the present study, a Missense polymorphism identified in the UCP gene at Ala118Val can be predicted to occur in the transmembrane region of the second domain close to the mitochondrial inner membrane space. The various biological functions of UCP are
![]()
Table 3. Association of the UCP genotypes at the growth and egg production traits (Mean ± S.E.).
a,bValues with different superscripts within the same row differ significantly (P < 0.05).
![]()
Table 4. Association of the UCP genotypes on breeding values of growth and egg production traits (Mean ± S.E.).
a,bValues with different superscripts within the same row differ significantly (P < 0.05)
not known in detail, and based on the localization and nature of the codon UCP118 variant, it is difficult to predict if the encoded protein change may cause defects in the functionality of the UCP. Structural-functional studies are thus needed to elaborate on the effect of the UCP Ala118Val variant on UCP’s protein structure as well as function. In a highly selected commercial population, changes in the allele and genotypic frequencies between generations may result from the net reproductive success of the individuals selected as breeding stock with the different genotypes and the selection norms adopted by the breeder. Variable associations of the identified polymorphisms may be a result of the differences in the population characteristics, sex, or both, indicating that the selection criteria may influence the production trait associations. This should be taken into consideration while selecting for the desired reproductive traits. Additional studies are required to expand the genetic, physiological, behavioral aspects involved in feed intake, digestion, and metabolism. The genomic diversity also has important implications in the evolutionary dynamics of species. Investigations of polymorphisms are useful for better understanding of the gene function, and those associated with commercially significant growth, breeding value of growth and reproductive traits have a potential for usage as molecular markers for selection programs. The identified polymorphisms and their associations with the traits of economic importance in present study provide greater insight into the role of gene involved in energy balance in poultry (UCP) and point toward the potential application of the findings for the enhancement of production traits by marker assisted selection.
Acknowledgements
The authors thank all the team who worked on this study in the experiments and by providing technical assistance in the laboratory.