The Use of Macroarray as a Simple Tool to Follow the Metabolic Profile of Lactobacillus plantarum during Fermentation ()
1. Introduction
Lactic acid bacteria (LAB) are widely used for the preservation of food and feed raw materials and to intensify the flavour and texture of fermented products. Of the lactobacilli commonly used in food processes, Lactobacil- lus plantarum is important in the production of many fermented foods of both plant (pickled vegetables, silage, sourdough) and animal (dry ferment sausages, fermented fish, cheese) origin [1] . This versatility and ecological flexibility is most likely associated with the genome size of L. plantarum, which is one of the largest known among LAB [2] . Based on complete genome sequencing, L. plantarum has a capacity to use a large variety of carbon sources and encompasses a relatively high number of regulatory functions concentrated within a defined genomic region, which was designated the lifestyle adaptation region [3] . These genomic features exhibit an ef- ficient adaptation capacity of L. plantarum to versatile environmental conditions.
Due to the ability to maintain pH homeostasis at low external pH, L. plantarum is tolerant to acidic environ- ment and often becomes the dominant LAB at the end of spontaneous vegetable fermentation [4] . Therefore, this species is common in vegetable and silage fermentations. Yet, spontaneous fermentation is generally poorly con- trolled and unstable, and the quality of products varies depending on fermented material and inherent microbiota. Spontaneously fermented vegetables may also contain among other things biogenic amines, which have been asso- ciated with certain toxicological characteristics and outbreaks of food poisoning. The formation of biogenic amines has been repressed by the use of a pure L. plantarum starter instead of spontaneous fermentation [5] . For the above reasons, well-characterized starter cultures with desirable properties would be of particular importance.
Previously, the technological properties of potential starter LAB could be determined almost exclusively in pilot- and full-scale food and feed production experiments. Today, the development of molecular techniques has made possible the exploitation of genomic and proteomic data for the observation of potential genotypic and phenotypic differences between individual strains in specific growth conditions. Studies based on L. plantarum DNA-microarrays [6] , proteomic patterns [7] - [10] and sequencing technologies, like metagenomic sequencing and RNAseq [11] - [13] and transcriptional profiling [14] - [17] have been carried out to elucidate strain-specific differences in genome composition and adaptation to various growth conditions.
Both microarray and proteomic studies require specific laboratory facilities and expertise, which may not be available in all research laboratories. Yet, L. plantarum is used worldwide for food and feed fermentations and there is a growing demand for the design of starter cultures with well-characterized technological properties. For easy and cost-efficient monitoring of gene expression in L. plantarum, a macroarray based on a group of se- lected L. plantarum genes printed onto a nitrocellulose filter was designed in this work. Using the macrofilters designed, the expression of a selected set of L. plantarum genes was assayed in synthetic MRS medium and in extracted carrot juice. To compare the potential differences of starter gene expression in hygienic and contami- nated cultivation media, the studied L. plantarum strain was cultivated in both sterile and contaminated (yeast and Escherichia coli) MRS and carrot juice.
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
L. plantarum strain MLBPL1 has been isolated from sauerkraut [18] [19] . The strain was routinely grown in mi- croaerophilic conditions at 32˚C and maintained in MRS broth (Difco, BD, Franklin Lakes, NJ, USA). For plat- ing, MRS was solidified with 1.5% agar. E. coli DH5a, carrying the plasmid vector pBluescript, was grown in Luria Bertani broth supplemented with ampicillin (50 µg/ml) as a selective agent at 37˚C 200 rpm. For con- tamination cultivations, a yeast and an E. coli strain originating from spoiled vegetables were propagated in YGC broth at 30˚C and in Luria Bertani broth at 37˚C, respectively.
For macroarray analyses, L. plantarum strain MLBPL1 was grown in synthetic medium MRS and carrot juice. To simulate contaminated growth conditions, the spoiling E. coli and yeast strains were inoculated into MRS and carrot juice. Cultivations were performed using Spectra/Por Float-A-Lyzer dialysis tube (MWCO 100 kDa, Spectrum Laboratories, Rancho Dominguez, CA, USA) in order to make it easier to separate the L. plantarum cells, the vegetable matrix and the spoiling strains of yeast and E. coli. By using the dialysis tube, no filtering of plant material was needed and in addition, the cells of contaminating strains didn’t interfere with the extraction of RNA. Carrot juice was prepared from fresh vegetables with a juice extractor. The extracted juice was centri- fuged at 18,500 g for 40 min and pasteurized in a water bath at 95˚C for 30 min.
An overnight culture of MLBPL1, grown in MRS-medium at 32˚C, was used to inoculate MRS broth and carrot juice. For MRS cultivation, a 1% inoculumn was used. For carrot juice cultivation and contamination cul- tivations, the inoculumn was centrifuged at 13,000 g for 3 min and the pellet was suspended into centrifuged (18,500 g for 40 min) and filter-sterilized (0.8/0.2 µm pore sizes) carrot juice (carrot cultivations) or MRS broth (MRS contamination cultivation), after which the suspension was transferred to a Spectra/Por Float-A-Lyzer dia- lysis tube. The tube was then transferred to a bottle containing carrot juice, contaminated MRS broth or con- taminated carrot juice. In contamination cultivations, MRS broth and carrot juice were contaminated by inocu- lating them with 1% E. coli and yeast. The cultivations were performed at 32˚C. The growth was determined by plating onto MRS agar plates appropriate dilutions from the samples taken during the growth. The plates were incubated at 32˚C for 48 h, until single bacterial colonies appeared.
2.2. Extraction and Labeling of RNA
Bacterial cells of the L. plantarum strain MLBPL1 grown in MRS broth and carrot juice were harvested at ex- ponential (6 h) and stationary phase (14 h) of growth by centrifugation for 3 min at 4˚C at 11,000 g. The col- lected cells were frozen immediately in liquid nitrogen and stored at −70˚C. Extraction of total RNA was carried out with SV total RNA isolation system (Promega, Madison, WI, USA) with some modifications to the protocol on the disruption of the cells. Briefly, bacterial cells thawed slowly on ice were first washed with sterile water treated with diethyl pyrocarbonate (DEPC) and collected by centrifugation. Next, the pellet was resuspended into 225 µl of SV RNA lysis buffer of the Promega kit and transferred to an eppendorf tube containing 100 µl of nitric acid-washed glass beads. The cells were disrupted with glass beads in a cell homogenizer as described be- fore (Kahala, et al., 2008). After that, the lysate was transferred to a new tube and 350 µl of SV RNA dilution buffer of the Promega kit was added per 175 µl of lysate. From this step on, the extraction was carried on as recommended by the manufacturer. Two technical duplicates from the RNA extraction on each culture medium and harvesting point were made. mRNA was enriched from total RNA samples by removing the 16S and 23S rRNAs with MICROB Express Bacterial mRNA Purification kit (Ambion, Austin, TX, USA) according to the instructions of the manufacturer. The RNA concentration was determined spectrophotometrically at 260 nm.
The integrity of the isolated prokaryotic RNA was determined by total RNA gel electrophoresis and Northern blot carried out as described by [20] . Total RNA samples, denatured with glyoxal and dimethylsulphoxide, were separated by size in a 1.0% (w/v) agarose gel in 10 mM sodium phosphate buffer, pH 6.5 followed by a transfer to a positively charged nylon membrane (Roche) and hybridization with a ldhD-specific 736 bp probe, amplified with primer pair 5’-AAGTTAGCCGACGAAGGG-3’ and 5’-CCATGTTGTGAACGGCAG-3’ targeted to L. plantarum strain D90339.1. The probe was labelled with digoxigenin-dUTP according to the instructions of the manufacturer (Roche). Luminescent DIG detection kit (Roche) was used for hybrid detection. To detect the po- tential residual chromosomal DNA in the isolated mRNA sample, primers 5’-AAGTTAGCCGACGAAGGG-3’ and 5’-GGGCGTATAATTCGTCCAAA-3’ designed to produce a 403 bp fragment from the target ldhD gene of L. plantarum strain D90339.1 were used. PCR-procedure using Dynazyme II DNA polymerase (Finnzymes, Espoo, Finland) was carried out in the reaction conditions recommended by the enzyme manufacturer.
cDNA was synthesized by RT from DNA-free mRNA and cDNA labelling was performed with an alkalilabile digoxigenin-11-dUTP (DIG) (Roche, Basel, Switzerland) in a reverse transcription reaction with Im-Prom-II Reverse Transcription System kit (Promega) as follows: 1 µg of mRNA was mixed with 0.5 µg of random hexamer primers provided by the kit manufacturer. The mixture was heated at 70˚C for 5 min and chilled on ice for 5 min. cDNA synthesis was carried out by combining RNA-primer mixture with 1 × ImProm-II reaction buffer, 5 mM MgCl2, 0.5 mM dATP, 0.5 mM dGTP, 0.5 mM dCTP, 0.325 mM dTTP, 0.175 mM DIG-11- dUTP, 1 U/µl RNasin Ribonuclease Inhibitor, and 1 µl ImProm-II Reverse Transcriptase. Annealing was per- formed at 25˚C for 5 min, followed by extension at 43˚C for 1h and enzyme inactivation at 70˚C for 15 min. The labeled cDNA was purified with Microarray Target Purification kit (Roche) according to the instructions of the manufacturer.
2.3. PCR and Labeling of the Positive Control for Macroarray
Human-based HbGAM (heparin-binding growth-associated molecule) gene [21] inserted into the plasmid pBluescript (Agilent Technologies, Santa Clara, CA) was amplified with polymerase chain reaction (PCR) to be used as a positive control in macroarray analyses. PCR reactions were carried out with Dynazyme II DNA po- lymerase (Finnzymes, Espoo, Finland) using the reaction conditions recommended by the manufacturer. Bacte- rial lysate of the E. coli strain DH5α, harbouring the recombinant pBluescript-HbGAM plasmid to be used as a template for PCR, was obtained by disrupting the cells with glass beads. To amplify the HbGAM gene, the pri- mer pair 5’-GTAAAACGACGGCCAG- 3’ and 5’-CAGGAAACAGCTATGAC-3’ targeting the plasmid was used. The amplified PCR product was purified with Wizard® SV Gel and PCR Clean-Up System (Promega) and labelled with DIG-11-dUTP using DIG-High Prime labeling kit (Roche).
2.4. Amplification of L. plantarum MLBPL1 Genes and Macroarray Printing
Primers were designed for the amplification of selected genes from the fully sequenced genome of L. plantarum WCFS1 [3] . The gene list and designed primers are listed in Supplement 1. To enable an easy re-amplification of the PCR-products, universal nucleotide sequences were added to the 5’ termini of the specific forward and reverse primers. A nucleotide sequence 5’ CCGCTGCTAGGCGCGCCGTG was added to the forward primers and, respectively, a nucleotide sequence 5’ GCAGGGATGCGGCCGCTGAC was added to the reverse primers. Amplification of the selected genes was done as described for the positive control for macroarray. For the PCR reaction, a 10 pmol primer concentration and 20 ng of corresponding genomic template DNA were used in a 100 µl reaction volume in a 96 well PCR plate. The success of the PCR amplification was checked by analyzing 5 µl from the reactions on a 1% agarose gel. The obtained PCR products were purified using Montage PCR Purifica- tion 96 Well Plates (Millipore).
PCR fragments in the 96 well plates were transferred to 384 plates for printing on the nitrocellulose macroar- ray (Supplement 2). Purified PCR fragments were gridded in duplicate on nitrocellulose membranes with a QPix automated colony picker (Genetix Ltd., UK) using a 384-pin gridding head as described in [22] .
2.5. Hybridization and Detection
Macroarrays were prehybridized for 2 h at 60˚C with 20 ml of DIG Easy Hyb buffer (Roche). Hybridizations were performed overnight at 60˚C with 6 ml DIG Easy Hyb buffer (Roche) containing 5 µl of labeled cDNA probe and HbGAM which was used as a positive control in hybridization reactions. After hybridization, macroarrays were washed twice at room temperature for 5 min with washing solution containing 2 × SSC (1 × SSC is 0.15 M NaCl and 15 mM sodium citrate) and 0.1% sodium dodecyl sulphate (SDS) and twice at 68˚C for 15 min with washing solution (0.1 × SSC, 0.1% SDS). Hybridized spots were detected with chemilumi- nesence-based DIG detection kit (Roche) using CDP-Star (Roche) as a substrate and chemiluminescence produced was detected by FluorChem (Alpha Innotech Corp., San Leandro, CA) gel image system. For re- probing, the DIG-labelled probe was removed with a following procedure. The membrane was rinsed thor- oughly in sterile water, washed twice with 0.2 M NaOH containing 0.1% SDS at 37˚C for 20 min and rinsed with 2 × SSC for 5 min.
2.6. Statistical Methods
The DNA probes spotted on the macroarray were selected using results from the previous proteomics results us- ing 2-DE and HPLC-ESI-MS/MS [8] . Additionally, computationally predicted expression values were used for choosing the remaining probes. Codon usage differences (codon bias) were used for predicting gene expression levels for all 3009 genes of L. plantarum (C) [3] , and the set of 63 genes encoding ribosomal proteins (RB). Codon bias for a gene g with respect to gene set G was calculated by the formula
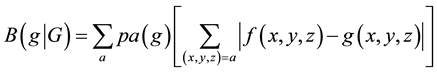 (1)
(1)
where f(x,y,z) denotes a normalized frequency of the codon triplet (x,y,z) coding for an amino acid a in a gene g, g(x,y,z) denotes the frequency of the codon triplet (x,y,z) in the gene set G, and pa(g) is the fraction of the amino acid a in the gene g [23] .
The gene g was predicted as highly expressed if the relative codon bias
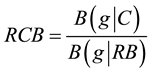 (2)
(2)
exceeded 1.05. The genes obtaining the greatest RCB values were chosen for the DNA macroarray filter in addi- tion to those identified by the HPLC-ESI-MS/MS.
After scanning of the macroarray images, the quantification of the hybridized signals and background sub- traction were done by the TIGR Spotfinder image processing software [24] . After quantification and background correction, the signals were normalized using median array intensities. Finally, the expression levels for each gene and sample was obtained by taking median of the normalized intensity values across the two replicates of each sample. Gene expression levels were compared between MRS, carrot juice and contaminated versions of the growth media in exponential (6 h) and stationary (14 h) growth phases. The pair-wise comparisons were made using fold changes, ratios of the mean expression levels. Fold changes greater than two are reported in the results. All genes in the array were grouped into functional groups according to their main roles. Gene set enrichment analysis was made for the gene sets with fold change greater than two in order to test whether any functional group is overrepresented among the differentially expressed genes between two growth media. Ana- lyses were made using SAS® (SAS for Windows 9.1).
3. Results
Growth rate of L. plantarum MLBPL1 cells was similar in MRS and carrot juice (Figure 1). Integrity and purity of the isolated RNA were demonstrated by Northern blot and PCR of the ldhD gene (data not shown).
The macroarray included 178 genes belonging to 18 main groups. The largest groups were energy metabolism (59 genes), protein synthesis (30 genes), protein fate (10 genes), regulatory functions (10 genes), cell envelope (9 genes) and DNA metabolism (9 genes). Of the 178 genes tested, 18 (10%) showed a mean fold change greater than 2.0 in at least one of the ten comparisons between growth media or growth phases (Table 1). The most frequent functions included were energy metabolism, cell envelope, protein fate and nucleotide meta- bolism.
3.1. Expression Levels of the Genes as a Function of Growth
The majority of the genes studied on the membranes showed no significant change in levels of expression during the growth or between the growth media. The number of the genes found to be regulated as a function of growth was clearly higher in MRS-based growth medium than in carrot juice, in which only genes involved in fatty acid and phospholipid metabolism showed differential expression in different growth phases. The function of the genes showing upregulation in logarithmic phase in MRS medium was mostly related to energy metabolism, but also to cell division, cell envelope biosynthesis and pyrimidine ribonucleotide biosynthesis. Generation of suffi- cient energy for growth in logarithmic phase is important and was evidenced in the MRS based medium.
In MRS cultivation, when entering in the stationary phase of growth, transcription of genes involved in energy metabolic pathways decreased and higher expression levels were found for genes involved in protein fate, pro- tein folding and stabilization, like “folding” chaperones DnaK and GroEL. In contaminated MRS medium, es- pecially the expression levels of genes involved in sugar metabolism pathways (galK, lacM) were found to be higher in logarithmic phase. This reflects higher demand for energy in the logarithmic phase and probably competition between the Lactobacillus and contaminating strains in the utilization of sugars that are needed for growth.
![]()
Figure 1. Growth of L. plantarum MLBPL1 in two growth media.
![]()
Table 1. Genes and their main roles showing mean fold change > 2.0 in ten comparisons among growth media and two growth phases. Minus and plus signs show which of the compared groups has higher expression: +: shows higher expression in the first; −: in the second group.
3.2. Expression Levels of the Genes between Different Growth Media
The mRNA level of several genes was shown to be regulated in response to different growth media. At the ex- ponential (6 h) phase of growth, the genes encoding dihydroorotate oxidase and dihydroorotase enzymes were differentially expressed in MRS and carrot juice. They showed 3.7- and 3.4-fold higher expression in the MRS compared to carrot juice growth medium, respectively (Table 1). Differential expression (p = 0.015) of these genes encoding proteins involved in pyrimidine ribonucleotide biosynthesis is an indication of distinct gene reg- ulation and, consequently, potentially different rate of pyrimidine biosynthesis in synthetic MRS compared to vegetable-based carrot juice cultivation medium.
Expression of malolactic enzyme (mle) gene was clearly higher in logarithmic phase when grown in plant based medium. Upregulation of cell division protein FtsH was observed in contaminated MRS 14 h compared to MRS 14 h, probably indicating higher stress response in contaminated MRS.
4. Discussion
This study focused on defining the differences in L. plantarum gene expression levels in different media and in different growth phases by the use of a simple and low-cost macroarray technique. Previously described DNA macroarray technique [22] has been further developed for studying gene expression profile of the industrially important lactic acid bacterium.
Fermentation conditions may dramatically affect functional characteristics of LAB [13] . Marked changes in expression levels upon entry in the stationary phase have been found out [25] [26] . Highly expressed genes are turned off or markedly repressed and genes, mostly inactive in the growing cells, begin to be expressed in the stationary phase [25] [26] . In this study, transcription of genes involved in energy metabolic pathways decreased in stationary phase and higher expression levels were found for genes like “folding” chaperones DnaK and GroEL. GroEL basal expression is enhanced by environmental stress, including elevated temperature, oxygen limitation, and nutrient deprivation [27] [28] . DnaK plays a central role in protein folding, refolding, transloca- tion and in the stress conditions. The elevated expression of these genes is probably a response to the diminish- ing nutrients and high concentration of lactic acid in the medium which is known to cause stress especially in the late-stationary phase [29] .
Proteomic studies by [10] has revealed significant changes on fermentation profiles of L. plantarum strains previously grown under food-like conditions compared to cultivation in MRS broth. In our study, expression of a malolactic enzyme (mle) gene in plant-based medium was found to be upregulated in logarithmic phase of growth. Mle enzymes, involved in decarboxylation of L-malic acid to L-lactic acid and CO2 [30] , have been pu- rified from several lactic acid bacteria, including Leuconostoc mesenteroides, L. plantarum, and Leuconostoc oenos [31] . In several studies, L. plantarum has been shown to have malolactic activity and therefore is of inter- est in wine production [32] . The significance of malolactic activity of LAB in sauerkraut fermentation has also been reported. Conversion of malic acid into lactic acid before significant sugar metabolism may play some role in early fermentation [30] [33] .
Higher expression of cell division protein FtsH in contaminated MRS compared to MRS probably indicated higher stress response in contaminated MRS. Functional studies have revealed an important role for FtsH in the bacterial stress response. In several bacteria, including E. coli, B. subtilis, Lactococcus lactis, O. oeni, Helico- bacter pylori, and L. plantarum, ftsH expression is induced in response to heat and other stress factors controlled by additional regulators [34] .
Macroarray was found to be an applicable method for studying expression of defined genes of L. plantarum during fermentation. Macroarray technology has been successfully applied also e.g. for the detection of patho- gens in chicken samples [35] and studies on environmental samples for the presence of specific antibiotic resis- tance genes [36] communities of diazotrophs [37] , and expression of 375 genes in L. lactis subsp. lactis IL1403 during stress conditions [38] .
L. plantarum is encountered in a variety of environmental niches, which include dairy, meat and many vege- table or plant fermentations as well as the human gastrointestinal tract. Because of this flexibility and versatility, strains of this species have been traditionally used for food and feed preservation and as starters in the manufac- ture of fermented products. Formerly, the technological properties and suitability of certain strains to selected applications could be ensured almost exclusively by laborious and time-consuming food processing and preser- vation experiments. Today, the long history of use and on the other hand the development of molecular and ge- nomic techniques have made L. plantarum one of the most studies food microbes. Modern DNA microarray [6] , next-generation sequencing technologies [39] and especially transcriptomic studies are accurate and sensitive and have enabled the detailed examination of L. plantarum genome structure and function. The most advanced technologies, however, require specific instrumentation and have often high running costs, which may rule out their use in many cases. The current study demonstrated that macroarrays printed on nitrocellulose filters with simple robotic systems can be analyzed by standard laboratory equipment and methods usually available in mo- lecular laboratories. Using this technology, rapid and cost-efficient analysis of genome function of L. plantarum can be carried out e.g. in developing regions, where lactic acid fermentation of food an feed matrices is a com- mon practice, but research and analysis laboratories often lack the most expensive specific laboratory instru- mentation.
Acknowledgements
Tekes, the Finnish Funding Agency for Technology and Innovation, is gratefully acknowledged for the financial support of this work. The authors wish to thank Anneli Paloposki for the skilful technical assistance, Ari-Matti Sarén for designing the primers, Markku Ala-Pantti and Hannu Väänänen for printing the membranes.