Natural Science
Vol.08 No.08(2016), Article ID:70235,21 pages
10.4236/ns.2016.88041
Reaction of Oxidized CuZnSOD with Polyphenols
Jack S. Summers1*, Benjamin Hickman1, Megan E. Arrington1, Bradley S. Stadelman2, Julia L. Brumaghim2, Michelle R. Yost1, Jeffrey D. Schmitt3, Mariah Hornby1, Stacy Sprague1
1Department of Chemistry and Physics, Western Carolina University, Cullowhee, NC, USA
2Department of Chemistry, Clemson University, Clemson, SC, USA
3The Bent Creek Institute, The North Carolina Arboretum, Asheville, NC, USA

Copyright © 2016 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 7 April 2016; accepted 28 August 2016; published 31 August 2016
ABSTRACT
Several polyphenolic compounds, including twelve flavonols and a variety of lower molecular weight compounds were found to diminish the effects of CuZnSOD on NMR relaxation of fluoride ion in a pH and concentration dependent manner. While we originally thought the effect arose from binding of the compounds to the enzyme active site, several lines of evidence indicated that active polyphenols reduced the paramagnetic copper(II) form of the enzyme to the diamagnetic copper(I) form, thereby giving false positive indications of enzyme inhibition in the NMR assay. First, docking experiments failed to provide a satisfactory explanation of the SAR. Second, effects on the enzyme’s EPR spectrum indicated that catechols could bind the active site copper leading to enzyme reduction. Third, while these reactions did not proceed to completion in aerobic solution, they did so under inert atmosphere. Fourth, experiments employing superoxide producing compounds demonstrated that loss of NMR activity did not prevent the enzyme from redox cycling. Thus, while the polyphenols appeared to inhibit the enzyme in the NMR assay, the compounds did not inhibit the enzyme’s reactions with superoxide.
Keywords:
Paramagnetic NMR, SOD, Polyphenol, Flavonol

1. Introduction
While inhibitors of superoxide dismutase enzymes (SODs) could be valuable tools for the study and management of oxidative stress, few compounds with this activity have been discovered. Based on the report that the steroid metabolite 2-methoxyestradiol (2ME) and related compounds inhibited CuZnSOD [1] - [3] , we began a study of the effects of planar polyphenolic compounds on the enzyme. While physiological effects of this compound were originally thought to arise from its effects on SOD activity, it was later shown that 2ME did not inhibit the enzyme [4] .
We began our study by looking at a number of flavonoids, a class of polyphenolic natural products. Flavonoids display both antioxidant and prooxidant activities [5] [6] that are associated with their widely ranging biological activities. Their antioxidant activities have variously been attributed to inhibition of xanthine oxidase [5] as well as metal sequestering [7] [8] mand radical scavenging [5] . Pro-oxidant activities have been attributed to the generation of reactive species (including superoxide) upon aerobic oxidation and to DNA damage by flavonoid/copper complexes [9] [10] . This latter activity has been proposed as a mechanism for their antibacterial and anticancer activities [11] . In addition to their redox related activities, flavonoids are known to bind to proteins [12] and to inhibit a variety of enzymes [13] , including ATPase [14] , topoisomerases [15] , integrases [16] and protein kinases [17] - [20] .
Conventional assays for SOD activity use redox-active chromaphores to monitor the fate of superoxide generated in situ by enzymatic or chemical means [21] . In the case of 2 ME, it was suggested that redox reactions of the chromaphore used in the assay caused the erroneous appearance of inhibitory activity. In addition, Huang et al. [1] and Soulere et al. [22] both reported that redox reactions of compounds from screening libraries with component(s) of the assay led to false results. The problem of false positives and false negatives arising from unaccounted side reactions of either superoxide or redox active chromaphores with compounds from the library under investigation must be expected for all assays that require the generation and/or use of chemically reactive substrates [1] [22] . Rapid disproportionation of superoxide in either neutral or acidic media also introduces com- plications in discovering SOD inhibitors.
Viglino et al. [23] and Rigo et al. [24] [25] demonstrated that CuZnSOD greatly accelerated the NMR relaxation of the fluoride ion 19F nucleus, and that the effect could be used to assay this enzyme [23] [26] . Recent papers by Bertini et al. [27] and Leung et al. [28] , describe use of proton relaxation (PRE) [29] methods to screen for compounds that bind to the active sites of other paramagnetic metalloenzymes. Because NMR methods use stable reporter nuclei to monitor accessibility of the metal ion at the enzyme active site, we reasoned that using these methods to screen for SOD inhibitors would circumvent complications associated with superoxide reactivity. NMR relaxation methods monitor the effects that paramagnetic metal ions at the enzyme active site have on spin coherence of observed nuclei in the NMR experiment. The SOD/fluoride interaction is especially sensitive and can be used to quantify CuZnSOD activity at enzyme concentrations as low as 10−7 M [24] . Since reaction of superoxide with the oxidized SOD is also believed to require contact with the active site metal ion [30] , we reason that any compound that binds the enzyme in a way that prevents access to the active site metal ion should prevent both 19F relaxation and also catalytic activity. We have reported that 19F NMR relaxation could be used to measure rates at which fluoride ion contacts the metal center in selected complexes of iron(III) and manganese(II), and that these rates correlate with the rates at which the complexes react with superoxide [31] , suggesting that these NMR methods could also be applied to discover compounds that prevent metal complexes from reacting with superoxide.
While the original objective of this work was to use NMR methods to search for inhibitors of CuZnSOD, redox reactions involving the enzyme and compounds under investigation led to several false positives. We believe that paramagnetic NMR relaxation can be used as a convenient measure of active site accessibility, but investigators must be careful that reactions that produce the diamagnetic reduced form of the enzyme do not interfere with the assay.
2. Experimental Section
2.1. General Methods
CuZnSOD from bovine erythrocytes was purchased from MP Biologicals. Flavonoids were purchased from Indofine Chemical Corporation. Buffers, NaF, trifluoroacetate (tfa), and other reagents were purchased from com- mercial sources and used without purification. 19F NMR spectra were acquired at 283 MHz (300 MHz 1H frequency) using a JEOL Eclipse NMR spectrometer. NMR solutions were prepared with 20 mM buffer, 20 mM NaF, 10% (v/v) D2O, and sufficient enzyme to give an easily measurable increase in transverse relaxation rate (R2 = 1/T2). Typically, the effects of potential inhibitors were measured under conditions where inhibitor-free, enzyme-containing control gave T2 values between 7 and 50 ms.
2.2. Compound Screening by NMR
Screening experiments employed a one-dimensional experiment using 2 mM trifluoroacetate (tfa) as an internal reference. While the one dimensional experiments employed the CPMG pulse sequence, spectra were acquired at a single relaxation delay. The 19F resonance integrals for fluoride and tfa were compared to determine the effects of potential inhibitors on the NMR relaxation activity of the enzyme.
2.3. EPR Spectroscopy
CuCl2 or CuZnSOD (50 µM), were allowed to incubate for known times with or without 10 mM polyphenol in 100 mM buffer, (either Tris, pH 8 or glycine/NaOH, pH 11) at room temperature. Samples without buffer were prepared using the same concentrations in deionized water, and the pH was adjusted by addition of NaOH to the appropriate value. Sample volumes were 0.2 mL. EPR spectra were measured at 120 K on a Bruker EMX spectrometer in quartz EPR tubes (0.3 mm i.d.). EPR samples were flash-frozen in liquid nitrogen and loaded into a pre-cooled cavity. Reported spectra are the average of five scans with a modulation amplitude of 10 G, modulation frequency of 100 kHz, microwave power of −1.0 mW, microwave frequency of 9.432 GHz, time constant of 40.96 ms, conversion time of 81.92 ms, and a sweep width of 1000 G centered at 3100 G. Spectra were corrected for residual cavity signal.
2.4. Metal Sequestering
A 2 mL sample containing approximately 4400 units of SOD (20 mM glycine buffer, pH 10) was divided into two aliquots and 16.2 μL of either DMSO or 123 mM methyl 3,4-dihydroxybenzoate (4, MDHB) solution in DMSO were added. The control and 4 containing solutions were allowed to equilibrate overnight at room temperature. Aliquots of the experimental and control solutions were removed for analysis of total copper and the high molecular weight components were removed from the remaining samples using YM-10 membrane filtration devices (10 KDa molecular weight cutoff, Millipore). Aliquots of the four samples were treated with aqueous HNO3 and copper concentrations were determined by Inductively Coupled Plasma Optical Emission Spectroscopy (ICP-OES) using a Perkin Elmer ICP-OES Optima 4100 DV spectrometer.
2.5. Characterizing Inhibition of NMR Activities
DMSO solutions with a range of polyphenol concentrations were prepared by serial dilution at 50 times their intended final concentration. NMR samples were prepared by addition of 12 µL aliquots of these DMSO solutions to 588 µL enzyme/buffer/fluoride solution. Thus, NMR solutions containing varied concentrations of polyphenol were prepared with a constant 2% (v/v) DMSO. Fluoride ion 19F transverse relaxation rates were determined by the standard CPMG method [32] . Fractional enzyme activities (A) in the NMR assay were calculated for inhibited solutions according to Equation (1);
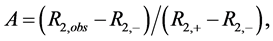 (1)
(1)
where R2,obs, R2,+ and R2,− are the transverse relaxation rates of the sample and controls containing either the same amount of enzyme or none, respectively. Inhibition profiles were determined by measuring fractional activities of samples as a function of polyphenol concentration.
2.6. Computational Experiments
The Molecular Operation Environment (MOE) program (Chemical Computing Group, Montreal) was used to dock nine flavonol structures to the active site of molecule 1 from the 1PU0 structure of CuZnSOD. Flavonolstructures were built and energy minimized in MOE as anions, deprotonated at the 4’ position. The enzyme structure was energy minimized after proton positions and partial charges had been assigned. Atoms within 12 Å of the copper atom were considered part of the active site. The flavonols were docked to the selected active site using the Triangle Matcher placement method. The docked poses were energy minimized using Force Field minimization and the energies of the poses were scored using the London DG method.
2.7. Effects of Oxygen on Inhibition of CuZnSOD NMR Relaxation by Polyphenols
Samples of enzyme in buffered solution were sparged with argon and aliquots of either MTHB or quercetin were added anaerobically. Using the NMR assay, samples were tested periodically for several hours, and again after overnight incubation.
3. Results and Discussion
3.1. Enhancement of Fluoride Ion 19F NMR Relaxation by CuZnSOD
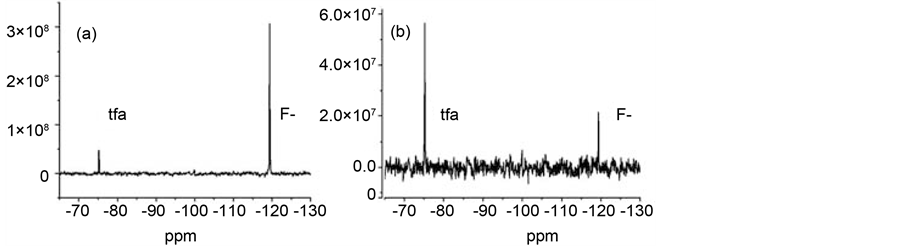
The effects of oxidized CuZnSOD on fluoride ion 19F NMR relaxation can be monitored using one-dimensional experiments if an internal reference is present in the assay solution. To illustrate, the effects of CuZnSOD on 19F NMR spectra of solutions containing F− and tfa are presented in Figure 1. Panel A in this figure shows the spectrum in the absence of enzyme, panel B shows the spectrum of the same solution but with ~10−7 M CuZnSOD added. Both spectra were acquired in one transient using a CPMG pulse sequence [32] with a 100 ms relaxation delay. These spectra demonstrate that SOD causes diminution of the peak intensity of the fluoride resonance relative to that of the tfa resonance. Inhibiting the NMR relaxation activity of the enzyme restores the intensity of the fluoride resonance.
3.2. Effects of Polyphenols on NMR Relaxation Enhancement by CuZnSOD
A variety of polyphenolic organic compounds were tested for their abilities to inhibit 19F NMR relaxation by CuZnSOD. In an initial screen of seven natural products (apigenin, biochanin a, coumestrol, coumestrol dimethyl ether, quercetin, genistein, and daidzein), only quercetin: 1) affected a diminution of the 19F NMR relaxation rate. We note that the loss of activity did not occur in the absence of 1, nor does it appear to result from non- specific interaction governed by hydrophobicity; Overnight incubation of enzyme with more hydrophobic compounds such as apigenin; 2) had no effect on the relaxation activity of the enzyme. Subsequently, we measured the effects of a series of related compounds, including a variety of flavonoid natural products and lower molecular weight species. The effects of the compounds on the NMR relaxation rate were strongly dependent on pH, molecular structure, compound concentration, and dissolved oxygen. Structures of these compounds and further details of their interactions with the enzyme in aerobic solution are presented in the Supplemental section of this manuscript.
3.3. The Effects of pH on Inhibition of NMR Activity
The effects of pH on the NMR activity were consistent with activity requiring deprotonation of a phenol group, 1 was less potent at pH 7 than at pH 8, and inhibitory activity dropped off even more significantly under acidic conditions. The pH dependence should not be surprising since flavonols typically have pKa values around 7, 9, and 11 [33] - [37] and precipitate from neutral or acidic solutions [15] . To further probe the pH dependence, we measured the effects of small molecules including benzoic acid and ester derivatives on NMR relaxation by the
Figure 1. Effect of SOD on 19F spectra. Panel (a): One transient CPMG spectrum of diamagnetic F− (20 mM NaF) and tfa (5 mM) solution at 282 MHz (100 ms relaxation; 300 MHz sectrometer). Panel (b): Effect of ~100 units CuZnSOD.
enzyme. Results of these experiments indicated that: 1) deprotonation of a carboxylic acid was not sufficient to induce inhibitory activity; 2) phenolic deprotonation is required; and 3) deprotonation of the enzyme at high pH renders it more susceptible to reaction. These results are described in greater detail in Supplemental Section 5.7.
3.4. The Effects of Polyphenol Structure on NMR Relaxation Activity of CuZnSOD
The effects of twelve flavonoids and nine additional low molecular weight compounds on CuZnSOD NMR activity in aerobic aqueous solution were measured at pH 8.0. Inhibition of the NMR activity under these conditions depended strongly on compound structure; flavonols having hydroxyl groups at the 3, 3’ and 4’ positions were more effective than lower molecular weight polyphenols or flavonoids lacking any of these groups. Representative results are shown in Figure 2 for inhibition of the NMR activity by the flavonol, myricetin (3).
3.5. The Effects of Polyphenol Concentration on CuZnSOD NMR Relaxation Activity
As shown in Figure 3, enzyme activity (calculated as described in the experimental section) was decidedly non-linear with polyphenol concentration. For many of the polyphenols, the behavior was consistent with aggregation limiting the availability of the active species. In most cases, the inhibitory behavior was well modeled by assuming a monomer/dimer equilibrium, with the monomer being the active species; The trend line in Figure 3 represents behavior expected for such a case. The concentration dependence of the fluorescence spectra of several flavonols supported this hypothesis. From the concentration dependence of the NMR activity inhibition data and the fluorescence data, it was possible to develop a structure/activity relationship for NMR activity inhibition by polyphenols. Inhibition of NMR activity by flavonols required hydroxyl groups at the 3, 3’ and 4’ positions. More detailed descriptions of our fluorescence results and of how NMR activities varied with compound structure are provided in the Supplemental Section.
3.6. Investigating the Possible Loss of Copper from SOD
Since the 3’ and 4’ hydroxyl groups constitute a catechol group on the flavonoid B ring, and since catechols (including 1 [38] ) form stable copper complexes [39] we considered the possibility that the catechol might chelate
Figure 2. Structures of representative polyphenols; quercetin (1), apigenin (2), myricetin (3), MDHB (4) and MTHB (5).
Figure 3. Effects of myricetin (3) concentration on 19F relaxation enhancement by CuZnSOD at pH 8.
the active site metal ion. Examples of species that inhibit CuZnSOD by interacting with the active site copper ion include low molecular weight anions (like cyanide and azide [40] - [44] ) and chelating agents like dithiocarbamates [45] - [48] . We note that catechol was among compounds reported to inhibit CuZnSOD by interacting with the active site copper [49] . The association equilibrium constant reported for copper binding by 1 (log(K) = 9.45 [50] ), however, is below the threshold reported for inhibition of SOD by metal binding (log(K) = 12.6) [49] .
While the low molecular weight CuZnSOD inhibitor diethyldithiocarbamate (DDC) acts by sequestering Cu2+ from the enzyme active site [48] , our results indicate that methyl-dihydroxybenzoate (MDHB, 4) does not act by this mechanism. In these experiments, we determined the concentrations of low molecular weight copper in enzyme samples after incubation in the presence and absence of 4. Copper concentrations were determined by inductively coupled plasma optical emission spectroscopy (ICP-OES). Low molecular weight fractions were isolated using Millipore YM-10 centrifugal concentration devices. Since the YM-10 device retains species larger than 10,000 Daltons, copper detected in the elutant should only arise from low molecular weight species. In these experiments, concentrations of copper in the low molecular weight fractions were not significantly affected by pre-incubated with the NMR inhibitor 4. In the 4 treated sample, 21% of the initial copper eluted as low molar mass material while 18% of copper in the control sample eluted. We conclude that the loss of NMR activity could not be attributed to the compound sequestering the metal from the enzyme.
3.7. Results of Computational Experiments
To evaluate the feasibility of an inhibition mechanism where the compounds bound the metal in the active site, we used the Molecular Operating Environment (MOE, Chemical Computing Group, Montreal, Canada) suite of programs to dock the flavonoids to the 1PU0 structure of CuZnSOD. While these experiments indicated that the active site could accommodate the flavonols, most poses generated by docking calculations did not present the flavonol in contact with the enzyme copper atom. While it was possible to generate the proposed active site by tethering the flavonol B ring to the copper via the 3’ and 4’ hydroxyl groups, energy minimization did not return significantly stabilized structures. Thus, the docking results did not help rationalize the observed structure activity relationship.
3.8. Results of EPR Studies
To investigate the effects of the compounds on the active site copper, we measured the EPR spectrum of the oxidized enzyme in the presence and absence of two low MW polyphenols (4, MDHB and 5, MTHB) at pH 8 and 11. Representative EPR spectra collected at pH 8 are presented in Figure 4. In the absence of polyphenol, EPR spectra of CuZnSOD are consistent with the reported spectra at both pH 8 and 11, (g = 2.28, A = 178 [51] ). As shown in Figure 4(a), addition of 10 mM 4 (MDHB) to the enzyme results in a spectrum with sharp features that resemble those in the spectrum of Cu2+-MDHB (prepared from CuCl2 and presented for comparison in the upper trace). Comparing the signal intensities in the spectra of CuZnSOD obtained before addition of 4 to that obtained 24 hours later, it appears that the total Cu2+ concentration is not grossly affected by the compound.
Figure 4. Panels (a) and (b) show the effects of MDHB (4) and MTHB (5) on the EPR spectrum of CuZnSOD at pH 8. For each panel, the lower trace shows the spectrum of the enzyme without added compound, the middle trace was recorded after 24 h incubation of enzyme with the polyphenol, and the upper trace shows the effect of the polyphenol on the spectrum of free copper(II) ion.
While the similarity of the spectrum to that of the Cu2+ complex of 4 suggests coordination, the centrifugal concentration experiments described above indicate that copper is not lost from SOD under these conditions. Thus, 4 does appear to coordinate copper in the active site of the protein.
In contrast to the effects of 4, addition of 10 mM 5 (MTHB) at pH 8 (Figure 4(b)) has less of an effect on the features of the spectrum but caused the resonance intensity to diminish with time. After 24 hour incubation with 10 mM 5, the Cu2+ resonance was nearly unobservable. We have verified that 5 reduces free Cu2+ at both pH 8 and 11 with loss of EPR signal. The increased ability of 5 (relative to 4) to reduce Cu2+ to Cu+ has been observed previously by EPR spectroscopy [52] . The difference in activities most likely reflects the difference in the redox potentials of the two compounds (Epa values for 4 and 5 are 0.380 and 0.293 V, respectively) [39] . At pH 11, addition of either 4 or 5 to CuZnSOD resulted in a diminution of the Cu2+ signal intensity with time and a yellowing of the solution, indicating oxidation of the polyphenol (data not presented) [53] - [56] . In the case of 5, a sharp feature consistent with the resonance of a semiquinone radical (g = 2.003 [56] ) was transiently observed in the EPR spectrum (data presented in Supplemental section, part 5.10.). Sharp features were also observed at frequencies consistent with resonances of the Cu2+ complex of 5 at this pH.
3.9. Effects of Oxygen on Inhibition of the NMR Activity
The effects of oxygen on inhibition of the NMR activity were studied using either 1 mM 5 (MTHB) or 50 µM 1 (quercetin) (concentrations where ~50% inhibition of the NMR activity is observed in aerobic solution). In contrast to the behavior in aerobic environment, overnight incubation of CuZnSOD with either 1 or 5 under inert atmosphere resulted in complete loss of NMR activity. The activity versus time profiles for these experiments is presented in Figure 5. Sparging the completely inhibited samples with O2 resulted in a nearly total restoration of NMR relaxation activity. These results are inconsistent with the possibilities that inhibition of the enzyme NMR activity involves reaction with oxygen radicals generated by aerobic oxidation of the organic compounds or with the products of such an oxidation.
3.10. Mechanism by Which Polyphenols Inhibit NMR Relaxation by CuZnSOD
Our results suggest that redox reactions (Equation (2) and Equation (3)) account for NMR activity inhibition.
 (2)
(2)
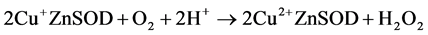 (3)
(3)
Copper(II) salts will oxidize catechols [57] and are known to catalyze their aerobic oxidation to quinones [58] . The redox hypothesis predicts that the degree of apparent inhibition induced by a given reducing agent in aerobic solution would reflect the steady state concentrations of oxidized and reduced enzyme. Since the reactions in Equation (2) and Equation (3) appear to proceed to completion, and since Equation (3) is independent of the identity of the polyphenol, the SAR for the aerobic inhibition of NMR activity should reflect differences in the forward rate constants for Equation (2) for the different flavonols. These rates, in turn, are expected to reflect the differences in redox potentials of the compounds. A log/log plot of equilibrium constants calculated from published redox potentials versus IC50 values from our NMR experiments (Figure 6) shows that the two parameters correlate. While a rigorous analysis would require substantially more data, the results presented in Figure 6 qualitatively support the hypothesis.
Figure 5. Under inert atmosphere, SOD NMR activity by either 1 mM 5 (MTHB) or 50 M 1 (quercetin).
Since the Cu+ form of the enzyme is an intermediate in the enzymatic reaction, the redox hypothesis predicts that reaction with the polyphenols would not affect the ability of the enzyme to catalyze superoxide dismutation. To test the redox competence of the inhibited enzyme, we measured the effect of 1 on the steady state concentration of oxidized CuZnSOD under redox cycling conditions. Rigo et al. [25] [26] , demonstrated that treating the oxidized enzyme with superoxide causes the NMR relaxation activity of CuZnSOD to decrease by about half, as a steady state mixture of the reduced and oxidized enzyme is established. The position of the steady state was reported to vary for different batches of the enzyme. We used a mixture of phenozine methosulfate (pms) and NADH to generate superoxide [59] and monitored its effect on fluoride relaxation by CuZnSOD. While 1 is known to react with superoxide, the rate constant for this reaction is much lower than that for reaction of superoxide with CuZnSOD (5 × 104 versus 2 × 109 M−1∙s−1, respectively) [60] [61] . Thus, the interference of 1 with the reactions of superoxide with the enzyme should be minimal. Results of this experiment are shown in Figure 7. The open triangles show the effects of superoxide on NMR relaxation by the enzyme in the absence of 1. The filled diamonds show the effects of 10 µM 1 on the reaction. The filled diamonds show that, prior to the addition of pms/NADH, the NMR relaxation activity of the 1 containing sample was ~67% that of the control. After addition of pms/NADH, the sample containing 1 lost relaxation activity, although at a lower rate than the control sample. Eventually the control and 1 containing samples reached the same relaxation rates. This result suggests that CuZnSOD forms the same steady state mixture of reduced and oxidized enzyme whether 1 is present in the solution or not. Thus, inhibition of the enzyme NMR relaxation activity by flavonoids appears to arise from reduction of the enzyme to give a product that is inactive in the NMR assay but retains its ability to catalyze superoxide dismutation.
4. Conclusion
In summary, we have demonstrated that enhancement of the fluoride ion 19F NMR relaxation rate by CuZnSOD can be assayed in one dimensional experiments using tfa as an internal reference. Using this technique, we have screened a variety of compounds for CuZnSOD inhibitory action and have discovered that flavonols inhibit the
Figure 6. IC50 values correlate with equilibrium constants calculated using flavonol oxidation potentials.
Figure 7. Effect of incubation with pms/NADH on 19F relaxation activity of CuZnSOD in the presence (filled diamonds) and absence (open triangles) of 1.
NMR relaxation activity of the enzyme. Our results indicate that inhibition of the NMR relaxation activity occurs via electron transfer. Interpretation of the inhibition data was complicated by aggregation of the inhibitors. Experiments with superoxide generating compounds indicated that reactions with the flavonoids did not prevent subsequent redox cycling of the enzyme.
Acknowledgements
We are grateful for support from the North Carolina Biotechnology Center (Basic Research Grant 556,603) and from the NIH (Academic Research Enhancement Award 1 R15 GM094034-01).
Cite this paper
Jack S. Summers,Benjamin Hickman,Megan E. Arrington,Bradley S. Stadelman,Julia L. Brumaghim,Michelle R. Yost,Jeffrey D. Schmitt,Mariah Hornby,Stacy Sprague, (2016) Reaction of Oxidized CuZnSOD with Polyphenols. Natural Science,08,359-379. doi: 10.4236/ns.2016.88041
References
- 1. Rigo, A., Stevanato, R., Viglino, P. and Rotilio, G. (1977) Competitive Inhibition of Cu, Zn Superoxide Dismutase by Monovalent Anions. Biochemical and Biophysical Research Communications, 79, 776-783.
http://dx.doi.org/10.1016/0006-291X(77)91179-2 - 2. Fee, J.A. and Gaber, B.P. (1972) Anion Binding to Bovine Erythrocyte Superoxide Dismutase: Evidence for Multiple Binding Sites with Qualitatively Different Properties. Journal of Biological Chemistry, 247, 60-65.
- 3. Bertini, I., Borghi, E., Luchinat, C. and Scozzafava, A. (1981) Binding Sites of Anions in Superoxide Dismutase. Journal of the American Chemical Society, 103, 7779-7783.
http://dx.doi.org/10.1021/ja00416a015 - 4. Banci, L., Bertini, I., Scozzafava, A. and Turano, P. (1989) Binding of Fluoride to Copper Zinc Superoxide Dismutase. Inorganic Chemistry, 28, 2377-2381.
http://dx.doi.org/10.1021/ic00311a024 - 5. Banci, L., Bencini, A., Bertini, I., Luchinat, C. and Piccioli, M. (1990) Hydrogen-1 NOE and Ligand Field Studies of Copper-Cobalt Superoxide Dismutase with Anions. Inorganic Chemistry, 29, 4867-4873.
http://dx.doi.org/10.1021/ic00349a011 - 6. Heikkila, R.E., Cabbat, F.S. and Cohen, G. (1976) In Vivo Inhibition of Superoxide Dismutase in Mice by Diethyl-dithiocarbamate. Journal of Biological Chemistry, 251, 2182-2185.
- 7. Heikkila, R.E., Cabbat, F.S. and Cohen, G. (1978) Inactivation of Superoxide Dismutase by Several Thiocarbamic Acid Derivatives. Experientia, 34, 1553-1554.
http://dx.doi.org/10.1007/BF02034668 - 8. Misra, H.P. (1979) Reaction of Copper-Zinc Superoxide Dismutase with Diethyldithiocarbamate. Journal of Biological Chemistry, 254, 11623-11628.
- 9. Cocco, D., Calabrese, L., Rigo, A., Argese, E. and Rotillio, G. (1981) Re-Examination of the Reaction of Diethyldithiocarbamate with the Copper of Superoxide Dismutase. Journal of Biological Chemistry, 256, 8983-8986.
- 10. Kelner, M.J., Bagnell, R., Hale, B. and Alexander, N.M. (1989) Inactivation of Intracellular Copper-Zinc Superoxide Dismutase by Copper Chelating Agents without Glutathione Depletion and Methemoglobin Formation. Free Radical Biology & Medicine, 6, 355-360.
http://dx.doi.org/10.1016/0891-5849(89)90079-8 - 11. Ni, Y., Du, S. and Kokot, S. (2007) Interaction between Quercetin-Copper(II) Complex and DNA with the Use of the Neutral Red Dye Fluorophor Probe. Analytica Chimica Acta, 584, 19-27.
http://dx.doi.org/10.1016/j.aca.2006.11.006 - 12. Malmstrom, B.G. and Vanngard, T. (1960) Electron Spin Resonance of Copper Proteins and Some Model Complexes. Journal of Molecular Biology, 2, 118-124.
http://dx.doi.org/10.1016/S0022-2836(60)80034-4 - 13. Perron, N.R., García, C.R., Pinzón, J.R., Chaur, M.N. and Brumaghim, J.L. (2011) Antioxidant and Prooxidant Effects of Polyphenol Compounds on Copper-Mediated DNA Damage. Journal of Inorganic Biochemistry, 105, 745-753.
http://dx.doi.org/10.1016/j.jinorgbio.2011.02.009 - 14. Kalyanaraman, B., Sealy, R.C. and Sivarajah, K. (1984) An Electron Spin Resonance Study of O-Semiquinones Formed during the Enzymatic and Autoxidation of Catechol Estrogens. Journal of Biological Chemistry, 259, 14018-14022.
- 15. Bors, W., Michel, C. and Stettmaier, K. (2000) Electron Paramagnetic Resonance Studies of Radical Species of Proanthocyanidins and Gallate Esters. Archives of Biochemistry and Biophysics, 374, 347-355.
http://dx.doi.org/10.1006/abbi.1999.1606 - 16. Seacat, A.M., Kuppusamy, P., Zweier, J.L. and Yager, J.D. (1997) ESR Identification of Free Radicals Formed from the Oxidation of Catechol Estrogens by Cu2+. Archives of Biochemistry and Biophysics, 347, 45-52.
http://dx.doi.org/10.1006/abbi.1997.0323 - 17. Perron, N.R. and Brumaghim, J.L. (2009) A Review of the Antioxidant Mechanisms of Polyphenol Compounds Related to Iron Binding. Cell Biochemistry and Biophysics, 53, 75-100.
http://dx.doi.org/10.1007/s12013-009-9043-x - 18. Kamau, P. and Jordon, R.B. (2002) Kinetic Study of the Oxidation of Catechol by Aqueous Copper(II). Inorganic Chemistry, 41, 3076-3083.
http://dx.doi.org/10.1021/ic010978c - 19. Balla, J., Kiss, T. and Jameson, R.F. (1992) Copper(II)-Catalyzed Oxidation of Catechol by Molecular Oxygen in Aqueous Solution. Inorganic Chemistry, 31, 58-62.
http://dx.doi.org/10.1021/ic00027a012 - 20. Rao, U.M. (1989) Superoxide Anion Radical-Independent Pathway for Reduction of Tetrazolium Salts in Aerobic Mixtures Consisting of NADH and 5-Methylphenazinium Methyl Sulfate in the Presence of Aqueous Micelles of Nonionic and Cationic Detergents. Free Radical Biology & Medicine, 7, 513-519.
http://dx.doi.org/10.1016/0891-5849(89)90024-5 - 21. Klug, D., Rabani, J. and Fridovich, I. (1972) A Direct Demonstration of the Catalytic Action of Superoxide Dismutase through the Use of Pulse Radiolysis. Journal of Biological Chemistry, 247, 4839-4842.
- 22. Jovanovic, S.V., Steeden, S., Tosic, M., Marjanovic, B. and Simicg, M.G. (1994) Flavonoids as Antioxidants. Journal of the American Chemical Society, 116, 4846-4851.
http://dx.doi.org/10.1021/ja00090a032 - 23. Argese, E., Viglino, P., Rotilio, G., Scarpa, M. and Rigo, A. (1987) Electrostatic Control of the Rate-Determining Step of the Copper, Zinc, Superoxide Dismutase Catalytic Reaction. Biochemistry, 26, 3224-3228.
http://dx.doi.org/10.1021/bi00385a043 - 24. Puerta, D.T., Lewis, J.A. and Cohen, S.M. (2004) New Beginnings for Matrix Metalloproteinase Inhibitors: Identification of High-Affinity Zinc-Binding Groups. Journal of the American Chemical Society, 126, 8388-8389.
http://dx.doi.org/10.1021/ja0485513 - 25. Clark, R.E.D. (1957) O-Dithiols in Analysis, Part III. Reactions of Toluene-3,4-Dithiol in Acetate Buffer and in Alkaline Solutions: Toluene-3,4-Dithiol as a Reagent for Copper, Cobalt, IronII, AntimonyV, and Thallium. Analyst, 82, 177-182.
http://dx.doi.org/10.1039/an9578200177 - 26. Hamilton, H.G. and Freiser, H. (1969) Extraction Equilibria for the System Toluene-3,4-Dithiol and Zinc. Analytical Chemistry, 41, 1310-1315.
http://dx.doi.org/10.1021/ac60279a007 - 27. Clark, R.E.D. (1958) O-Dithiols in Analysis, Part VII. Toluene-3,4-Dithiol as a General Analytical Reagent in Qualitative Analysis. Analyst, 83, 396-402.
http://dx.doi.org/10.1039/an9588300396 - 28. Clark, R.E.D. (1957) O-Dithiols in Analysis, Part IV. Diacetyltoluene-3,4-Dithiol, Dibenzoyltoluene-3,4-Dithiol and the Zinc Complex of Toluene-3,4-Dithiol. Analyst, 82, 182-185.
http://dx.doi.org/10.1039/an9578200182 - 29. Perron, N.R., Hodges, J.N., Jenkins, M. and Brumaghim, J.L. (2008) Predicting How Polyphenol Antioxidants Prevent DNA Damage by Binding to Iron. Inorganic Chemistry, 47, 6153-6161.
http://dx.doi.org/10.1021/ic7022727 - 30. Brown, J.E., Khodr, H., Hider, R.C. and Rice-Evans, C.A. (1998) Structural Dependence of Flavonoid Interactions with Cu2+ ions: Implications for Their Antioxidant Properties. Biochemical Journal, 330, 1173-1178.
http://dx.doi.org/10.1042/bj3301173 - 31. Zenkevich, I.G. and Guschina, S.V. (2010) Determination of Dissociation Constants of Species Oxidizable in Aqueous Solution by Air Oxygen on an Example of Quercetin. Journal of Analytical Chemistry, 65, 371-375.
http://dx.doi.org/10.1134/S1061934810040064 - 32. Sauerwald, N., Schwenk, M., Polster, J. and Bengsch, E. (1998) Spectrometric pK Determination of Daphnetin, Chlorogenic Acid and Quercetin. Zeitschrift für Naturforschung, 53, 315-321.
http://dx.doi.org/10.1515/znb-1998-0310 - 33. Musialik, M., Kuzmicz, R., Pawlowski, T.S. and Litwinienko, G. (2009) Acidity of Hydroxyl Groups: An Overlooked Influence on Antiradical Properties of Flavonoids. The Journal of Organic Chemistry, 74, 2699-2709.
http://dx.doi.org/10.1021/jo802716v - 34. Favaro, G., Clementi, C., Ramani, A. and Vickackaite, V. (2007) Acidichromism and Ionochromism of Luteolin and Apigenin, the Main Components of the Naturally Occurring Yellow Weld: A Spectrophotometric and Fluorimetric Study. Journal of Fluorescence, 17, 707-714.
http://dx.doi.org/10.1007/s10895-007-0222-0 - 35. Herrero-Martinez, J.M., Sanmartin, M., Roses, M., Bosch, E. and Rafols, C. (2005) Determination of Dissociation Constants of Flavonoids by Capillary Electrophoresis. Electrophoresis, 26, 1886-1895.
http://dx.doi.org/10.1002/elps.200410258 - 36. Braun, S., Kalinowski, H.-O. and Berger, S. (1998)150 and More Basic NMR Experiments. Wiley-VCH Weinheim, 159.
- 37. Summers, J.S., Baker, J.B., Meyerstein, D., Mizrahi, A., Zilbermann, I., Cohen, H., Wilson, C.M. and Jones, J.R. (2008) Measured Rates of Fluoride/Metal Association Correlate with Rates of Superoxide/Metal Reactions for FeIIIEDTA-(H2O)-and Related Complexes. American Chemical Society, 130, 1727-1734.
http://dx.doi.org/10.1021/ja077193b - 38. Valentine, J.S. (2007) Biological Inorganic Chemistry: Structure and Reactivity. In: Gray, H., Stiefel, E.I., Valentine, J.S. and Bertini, I., Eds., Biological Inorganic Chemistry: Structure and Reactivity, Chapter 3, University Science Books, Mill Valley, 331.
- 39. Dwek, R.A. (1973) Nuclear Magnetic Resonance in Biochemistry. Clarendon Press, Oxford, 247.
- 40. Leung, I.K., Flashman, E., Yeoh, K.K., Schoefield, C.J. and Claridge, T.D.W. (2010) Using NMR Solvent Water Relaxation to Investigate Metalloenzyme-Ligand Binding Interactions. Journal of Medicinal Chemistry, 53, 867-875.
http://dx.doi.org/10.1021/jm901537q - 41. Bertini, I., Fragai, M., Luchinat, C. and Talluri, E. (2008) Water-Based Ligand Screening for Paramagnetic Metalloproteins. Angewandte Chemie, 47, 4533-4537.
http://dx.doi.org/10.1002/anie.200800327 - 42. Viglino, P., Rigo, A., Argese, E., Calabrese, L., Cocco, D. and Rotilio, G. (1981) 19F Relaxation as a Probe of the Oxidation State of Cu, Zn Superoxide Dismutase. Studies of the Enzyme in Steady-State Turnover. Biochemical and Biophysical Research Communications, 100, 125-130.
http://dx.doi.org/10.1016/S0006-291X(81)80072-1 - 43. Rigo, A., Ugo, P., Viglino, P. and Rotilio, G. (1981) 19F-Nuclear Magnetic Relaxation by Superoxide Dismutase as an Enzymic Method for the Detection of Superoxide Ion. FEBS Letters, 132, 78-80.
http://dx.doi.org/10.1016/0014-5793(81)80431-0 - 44. Rigo, A., Viglino, P., Argese, E., Terenzi, M. and Rotilio, G. (1979) Nuclear Magnetic Relaxation of 19F as a Novel Assay Method of Superoxide Dismutase. Journal of Biological Chemistry, 254, 1759-1760.
- 45. Viglino, P., Rigo, A., Stevanato, R., Ranieri, G.A., Rotilio, G. and Calabrese, L. (1979) The Binding of Fluoride Ion to Bovine Cuprozinc Superoxide Dismutase as Studied by 19F magnetic Relaxation. Journal of Magnetic Resonance, 34, 265-274.
http://dx.doi.org/10.1016/0022-2364(79)90002-7 - 46. Soulere, L., Delplace, P., Davioud-Charvet, E., Py, S., Sergheraert, C., Perie, J., Ricard, I., Hoffmann, P. and Dive, D. (2003) Screening of Plasmodium falciparum Iron Superoxide Dismutase Inhibitors and Accuracy of the SOD-Assays. Bioorganic & Medicinal Chemistry, 11, 4941-4944.
http://dx.doi.org/10.1016/j.bmc.2003.09.011 - 47. Ewing, J.F. and Janero, D.R. (2005) Microplate Superoxide Dismutase Assay Employing a Nonenzymatic Superoxide Generator. Analytical Biochemistry, 232, 243-248.
http://dx.doi.org/10.1006/abio.1995.0014 - 48. Akiyama, T., Ishida, J., Nakagawa, S., Ogawara, H., Watanabe, S., Itoh, N., Shibuya, M. and Fukami, Y. (1987) Genistein, a Specific Inhibitor of Tyrosine-Specific Protein Kinases. Journal of Biological Chemistry, 262, 5592-5595.
- 49. McGovern, S.L., Caselli, E., Grigorieff, N. and Shoichet, B.K. (2002) A Common Mechanism Underlying Promiscuous Inhibitors from Virtual and High-Throughput Screening. Journal of Medicinal Chemistry, 45, 1712-1722.
http://dx.doi.org/10.1021/jm010533y - 50. Davies, S.P., Reddy, H., Caivano, M. and Cohen, P. (2000) Specificity and Mechanism of Action of Some Commonly Used Protein Kinase Inhibitors. Biochemical Journal, 351, 95-105.
http://dx.doi.org/10.1042/bj3510095 - 51. Matter, W.F., Brown, R.F. and Vlahos, C.J. (1992) The Inhibition of Phosphatidylinositol 3-Kinase by Quercetin and Analogs. Biochemical and Biophysical Research Communications, 186, 624-631.
http://dx.doi.org/10.1016/0006-291X(92)90792-J - 52. Fesen, M.R., Kohn, K.W., Leteurtre, F. and Pommier, Y. (1993) Inhibitors of Human Immunodeficiency Virus Integrase. Proceedings of the National Academy of Sciences of the United States of America, 90, 2399-2403.
http://dx.doi.org/10.1073/pnas.90.6.2399 - 53. Webb, M.R. and Ebeler, S.E. (2004) Comparative Analysis of Topoisomerase IB Inhibition and DNA Intercalation by Flavonoids and Similar Compounds: Structural Determinates of Activity. Biochemical Journal, 384, 527-541.
http://dx.doi.org/10.1042/BJ20040474 - 54. Lang, D.R. and Racker, E. (1974) Effects of Quercetin and F1 Inhibitor on Mitochondrial ATPase and Energy-Linked Reactions in Submitochondrial Particles. Biochimica et Biophysica Acta, 333, 180-186.
http://dx.doi.org/10.1016/0005-2728(74)90002-4 - 55. Teillet, F., Boumendjel, A., Boutonnat, J. and Ronot, X. (2008) Flavonoids as RTK Inhibitors and Potential Anticancer Agents. Medicinal Research Reviews, 28, 715-745.
http://dx.doi.org/10.1002/med.20122 - 56. Dangles, O., DuFour, C., Manach, C., Morand, C. and Remsey, C. (2001) Binding of Flavoniods to Plasma Proteins. In: Packer, L. and Sies, L., Eds., Bioflavonoids and Polyphenols, Academic Press, Pittsburgh, 319-333.
http://dx.doi.org/10.1016/S0076-6879(01)35254-0 - 57. Hadi, S.M., Bhat, S.H., Azmi, A.S., Hanif, S., Shamim, U. and Ullah, M.F. (2007) Oxidative Breakage of Cellular DNA by Plant Polyphenols: A Putative Mechanism for Anticancer Properties. Seminars in Cancer Biology, 17, 370-376.
http://dx.doi.org/10.1016/j.semcancer.2007.04.002 - 58. Bhat, S.H., Azmi, A.S. and Hadi, S.M. (2007) Prooxidant DNA Breakage Induced by Caffeic Acid in Human Peripheral Lymphocytes: Involvement of Endogenous Copper and a Putative Mechanism for Anticancer Properties. Toxicology and Applied Pharmacology, 218, 249-255.
http://dx.doi.org/10.1016/j.taap.2006.11.022 - 59. Hanif, S., Shamim, U., Ullah, M.F., Azmi, A.S., Bhat, S.H. and Hadi, S.M. (2008) The Anthocyanidin Delphinidin Mobilizes Endogenous Copper Ions from Human Lymphocytes Leading to Oxidative Degradation of Cellular DNA. Toxicology, 249, 19-25.
http://dx.doi.org/10.1016/j.tox.2008.03.024 - 60. Van Acker, S.A.B.E., Van Den Berg, D.-J., Tromp, M.N.J.L., Griffioen, D.H., Van Bennekom, W.P., Van Der Vijgh, W.J.F. and AaltBast, A. (1996) Structural Aspects of Antioxidant Activity of Flavonoids. Free Radical Biology & Medicine, 20, 331-342.
http://dx.doi.org/10.1016/0891-5849(95)02047-0 - 61. Perron, N.R. and Brumaghim, J.L. (2009) A Review of the Antioxidant Mechanisms of Polyphenol Compounds Related to Iron Binding. Cell Biochemistry and Biophysics, 53, 75-100.
http://dx.doi.org/10.1007/s12013-009-9043-x - 62. Sakihama, Y., Cohen M.F., Grace S.C. and Yamasaki, H. (2002) Plant Phenolic Antioxidant and Prooxidant Activities: Phenolics-Induced Oxidative Damage Mediated by Metals in Plants. Toxicology, 177, 67-80.
http://dx.doi.org/10.1016/S0300-483X(02)00196-8 - 63. Cotelle, N. (2001) Role of Flavonoids in Oxidative Stress. Current Topics in Medicinal Chemistry, 1, 569-590.
http://dx.doi.org/10.2174/1568026013394750 - 64. Kachadourian, R., Liochev, S.I., Cabelli, D.E., Patel, M.N., Fridovich, I. and Day, B.J. (2001) 2-Methoxyestradiol Does Not Inhibit Superoxide Dismutase. Archives of Biochemistry and Biophysics, 392, 349-353.
http://dx.doi.org/10.1006/abbi.2001.2455 - 65. Wood, L., Leese, M.R., LeBlond B., Woo, L.W., Ganeshapillai, D., Purohit, A., Reed, M.J., Potter, B.V. and Packham, G. (2001) Inhibition of Superoxide Dismutase by 2-Methoxyoestradiol Analogues and Oestrogen Derivatives: Structure-Activity Relationships. Anti-Cancer Drug Design, 16, 209-215.
- 66. Agoston, G.E., Shah, J.H., LaVallee, T.M., Zhan, X., Pribluda, V.S. and Treston, A.M. (2007) Synthesis and Structure-Activity Relationships of 16-Modified Analogs of 2-Methoxyestradiol. Bioorganic & Medicinal Chemistry, 15, 7524-7537.
http://dx.doi.org/10.1016/j.bmc.2007.09.011 - 67. Huang, P., Feng, L., Oldham, E.A., Keating, M.J. and Plunkett, W. (2000) Superoxide Dismutase as a Target for the Selective Killing of Cancer Cells. Nature, 407, 390-395.
http://dx.doi.org/10.1038/35030140
Appendix 1
1.1. Supplemental Section (Reaction of Oxidized CuZnSOD with Poly-Phenols, Summers, et al.)
1.1.1. Inhibition of the NMR Relaxation Activity of CuZnSOD by Flavonoids in Aerobic Solution
Interpretation of inhibition constants (Kinh); The effects of flavonols and lower molecular weight compounds on NMR relaxation enhancement by CuZnSOD were measured as described in the Experimental section of the manuscript. Normalized activities of inhibitor containing solutions were calculated as;
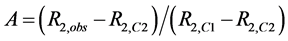 , where
, where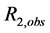 ,
, 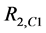 , and
, and  are the transverse relaxation rates of the observed sample and of a positive control containing enzyme but no inhibitor and negative control containing neither enzyme nor inhibitor, respectively. Data were originally treated as if inhibition resulted solely from binding of the polyphenols to the enzyme active site (Equation (S1)).
are the transverse relaxation rates of the observed sample and of a positive control containing enzyme but no inhibitor and negative control containing neither enzyme nor inhibitor, respectively. Data were originally treated as if inhibition resulted solely from binding of the polyphenols to the enzyme active site (Equation (S1)).
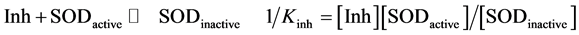
Our work (described in the Results and Discussion section), however, subsequently showed that NMR inhibition likely results from both binding to the enzyme and from electron transfer to the paramagnetic copper, with the latter probably giving the more important contribution to the most active compounds. If, as we propose, the relaxation activity of the enzyme is determined by steady state ratio of oxidized to reduced enzyme in aerobic solution is determined by the rates of the reactions in Equation (S2) and Equation (S3), then differences between apparent inhibition constants should reflect differences in the rates of reaction (S2) (Equation (S4)):
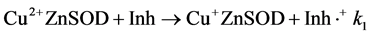
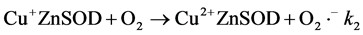
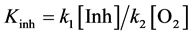
Chemical structures of the flavonoids used in this study are presented in Figure 8.
Figure 8. Structures of selected flavonoids.
1.1.2. Flavonol Aggregation Equilibrium Constants
As discussed in the Results and Discussion section of this manuscript, plots of 1/A-1 were distinctly non-linear with polyphenol concentration. We attribute this to dimerization and higher order aggregation of the compounds in aqueous solution. Equation (S1), above describes how the inhibitory activities of flavonoids could be affected by dimerization, this is a simplified scenario and the fluorescence data indicate that higher order aggregation occurs for some compounds. Characterizing such behavior requires consideration of trimerization, characterized by an equilibrium constant (Ktrimer, defined in Equation (S5)). In the general case, the total mass of material must be distributed between monomer, dimer, trimer, etc., as in Equation (S6). While a fuller description of aggregation would require an equilibrium constant for each multimer considered, our data could be modeled adequately assuming that only monomer, dimer and trimer were present in significant concentrations. For example, more than 98.8% of the variance in the fluorescence spectra of solutions containing from 1 to 512 M 3 could be accounted for by the three species.

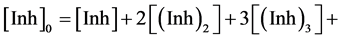

A complete description of the behavior predicted by the involvement of trimers would require solution of a third order polynomial. While roots of the cubic equation can be calculated, the exact solution does not lend itself to least squares refinement since solutions having an imaginary component can arise under different scenarios. To simplify the calculation, we made two simplifying assumptions. First, we limited our model by assuming 1/Ktrimer would always be greater than or equal to 1/Kdimer. Since the individual units all carry a negative electronic charge, it is reasonable that electrostatic repulsion will destabilize larger aggregates relative to smaller ones. Our second assumption was that the concentration of monomer could be approximated by that calculated in the absence of trimerization (the dimer only case). The validity of this assumption can be assessed by comparing calculated distribution information for different assumed values of Ktrimer. Figure 9 shows the fraction of monomer for a system with 1/Kdimer = 20 M assuming different values for 1/Ktrimer. The dashed line represents the dimerization only case, the solid blue and black lines represent the cases where 1/Ktrimer = 20 and 100 M, respectively. Comparing the solid blue line to the dashed line shows that even in the worst case (when dimerization and trimerization constants are equal), the effect does not grossly distort the concentration profile. As shown by comparison of the solid black and dashed lines, the fit improves with an increasing difference in the two constants.

In the dimerization only case, Equation (S7) gives a quadratic and the concentration of monomer can be estimated using Equation (S8). The remaining material can be partitioned between dimer and trimer using Equation
Figure 9. Distribution diagram for monomer concentration for the case where 1/Kdimer = 20. The dashed black line represents the fraction of monomer for the case of where no trimerization occurs. The solid blue and black lines present the behavior for 1/Ktrimer = 20 and 100 M respectively.
(S5) and Equation (S6). Modeling the data in this way allowed us to approximate concentrations of monomer, dimer, and trimer for any total inhibitor concentration given an assumed combination of equilibrium constants. Molar fluorescence coefficients for the monomer, dimer, and trimer were then estimated by least squares optimization to minimize the difference between predicted and measured spectra. To illustrate, we will consider two cases, one where trimerization was not important at low concentrations (myricetin, 3) and the other where trimerization was important (gossypin, 6). The observed and predicted fluorescence data for 6 are presented in Figure 10. Panels A and B in this figure show the measured and calculated emission spectra, while panel C shows the sensitivity of the fit (as the sum of the squares of the residuals) to changes in 1/Kdimer and 1/Ktrimer. As shown in panel C, the best fit of the data was achieved when 1/Kdimer was between 5 and 30 M and 1/Ktrimer was between 40 and 60 M. While uncertainties would be best considered on a log scale, we chose to report these using the more accepted linear scale.
In contrast to the behavior of 6, 3 appeared to only form significant amounts of trimer at the highest concentrations where fluorescence spectra were recorded. The effects of concentration on the emission spectrum of 3 are presented in Figure 3 of the manuscript and in Figure 11 and Figure 12, here. As noted in the manuscript, the normalized fluorescence spectra of the 3 at low concentrations displayed isosbestic behavior within experimental uncertainty. Figure 10 shows the normalized fluorescence spectrum of myricetin ranging from 1 to 64 M with expanded regions surrounding the four isosbestic points. Uncertainties in fluorescence intensities can be gauged from noise levels in the spectra. We conclude that only two major 3-containing species are present between 1 and 64 M. When fluorescence data for all measured wavelengths are considered, the best fit is found at 1/Kdimer = 24 M. From the plot of the SSR parameter versus the assumed 1/Kdimer, we estimate the uncertainty of this value as ~5 M.
Figure 10. Observed and modeled emission spectra of 6. (a) Measured spectra recorded at concentrations ranging from 1 to 512 mM. (b) Spectra calculated using least square minimization. (c) Sensitivity of the sum of the squares of residuals to changes in Kdimer at three values of Ktrimer.
Figure 11. (a) Normalized fluorescence spectra of myricetin at concentrations between 1 and 64 M. Panels (b) and (c) show expanded regions displaying isosbestic like behavior.
Figure 12. Inhibition of CuZnSOD by myricetin (3). Data from two independent experiments. Trend lines show behavior predicted for cases where 1/Kdimer = 20 M and 1/Kinh = 5.3, 6.0, and 6.6 M. Error bars assume 10% uncertainty in (1/A-1) and 7% uncertainty in concentration.
Uncertainties in 1/Ktrimer were too great to justify reporting their values. Even when we have evidence for significant trimerization at lower concentrations, our fitting parameter (SSR) was relatively insensitive to changes in this parameter. As shown in Panel C of Figure 11, SSR curves overlapped substantially when 1/Ktrimer was between 40 and 60 M. Further, evidence against higher order aggregation is not sufficiently compelling to argue that deviation arose from trimerization alone.
1.1.3. Experimental Uncertainties
Uncertainties in the results of NMR experiments arise from a number of factors. For these experiments, samples were prepared by a series of steps. The mass of solid inhibitor was measured to ~1%. The solid sample was dissolved in a volume of DMSO that is known to ~2%. DMSO solutions of descending concentration were prepared by serial dilution. NMR solutions were prepared by dilution of one of the DMSO solutions in enzyme containing buffer. If each dilution added another 2% uncertainty, then the highest concentration NMR sample should have an inhibitor concentration known to approximately 7%, and each subsequent sample is known to 2% lower accuracy. Thus, accumulation of errors leads the concentrations of more dilute solutions to be less certain than those of more concentrated samples. Measured relaxation rates (R2 values) were reproducible to within ~3% on a given sample and activities were determined from measurements of three samples (the experiment and two controls). Since the 1/A-1 value is the ratio of two differences (Equation (1), Experimental Section), the greatest accuracy is achieved at unity with uncertainty increasing at both larger and smaller values. In the worst case, we estimate that 1/A-1 values should be good to within 10%. Experimental reproducibility can be gauged by comparing the results of independent measurements of inhibition by the same compound. Figure 11 shows inhibition curves for two measurements using 3. Error bars in this figure show 10% uncertainty in 1/A-1 and 7% in concentration.
1.1.4. Uncertainties in Kinh Values
Using limits on 1/Kdimer established by the fluorescence experiments (20 and 40 M), it is possible to estimate limits on acceptable values of Kinh. The dependence of enzyme activity on total added inhibitor comes from substituting the monomer concentration from the fluorescence experiments into Equation (S1). Trend lines in Figure 11 represent behaviors predicted for 1/Kdimer = 20 M and 1/Kinh values of 5.3, 6.0, and 6.6 M. The two outer values represent the limiting values where the error bars on the 80 M data point still contain the projected value. A similar analysis conducted using 1/Kdimer = 40 M gave a best fit value for 1/Kinh of 7.7 M with limits of 6.9 and 8.4 M. This analysis leads us to report values of Kdimer and 1/Kinh for 3 as 24 ± 6 and 6.8 ± 1.6 M, respectively. Values and uncertainties in Kdimer and 1/Kinh for the other inhibitors were determined similarly.
1.1.5. Structure/Activity Relationship for Inhibition of the NMR Relaxation Activity of CuZnSOD by Flavonoids
Comparing the 1/Kinh values in Table 1 shows that having hydroxyl groups at the 3, 3’, 4’ and 5’ positions enhance inhibition. The effects of hydroxyl substitution at the 3 position can best be seen by comparing the activity
Table 1. CuZnSOD inhibitory and dimerization data for selected flavonoids at pH 8.0.
Notes: a) 1/Kdimer values should be accurate to within a factor of three. b) 1/Kinh values should be accurate to within a factor of two c) Half wave potentials for flavonoid oxidation, adjusted for comparison to the normal hydrogen electrode, NHE. Values are from reference S7. d) Kdimer could not be determined accurately from fluorescence data. e) IC50 estimated by extrapolation of reciprocal activity versus [Inh]0.5 plot.
of 1 (1/Kinh = 24 M) to that of 13 (1/Kinh > 200 M). These two compounds only differ at the 3 position where 1 has the hydroxyl group and 13 has hydrogen. We considered the possibility that the importance of the 3-hydroxyl group to inhibition arises from its ability to tautomerize to give a ketone at this position. To test this hypothesis, we measured the effect of 9 on the enzyme. The structure of 9 differs from that of 1 in that the bond between the 2 and 3 position carbon atoms is a single bond in 9 whereas it is a double bond in 1. Thus, the 3 hydroxyl group on 1 may tautomerize to give a ketone while 9 cannot. Comparing IC50 values for the compounds in Table 1 shows that inhibition by a mixture of 9 isomers was similar to that displayed by other active flavonoids. We conclude that tautomerization of the 3 position hydroxyl is not critical to inhibition.
It is apparent that compounds containing the catechol moiety on the B ring are more effective inhibitors than are compounds that do not. For example, 10 (having a single B ring hydroxyl group) is much less effective against CuZnSOD than 1 (having B ring hydroxyl groups at the 3’ and 4’ positions). Further, the importance of the catechol can be seen by comparing the inhibitory activities of two isomeric flavonols, 12 and 1. The structures of the compounds differ in the positions of the hydroxyl groups on the B ring. While 1 has hydroxyl groups on the 3’ and 4’ positions, the 12 hydroxyls are at the 2’ and 4’ positions. As was the case with 10, 12 is also a poor inhibitor.
In contrast to the dramatic effects observed when substitution patterns on the B ring are changed, the number and positions of hydroxyl groups on the A ring had a lesser impact on inhibition. We measured the inhibitory activities of five active compounds that only differ in A ring substitution patterns (1, 6, 7, 8, 11). While IC50 values (reported in Table 1) varied by up to a factor of 10, further analysis indicated that the differences arose primarily from differences in the flavonol dimerization constants. Further, comparing the activities of 1 and 6 indicates that incorporation of a bulky glucose moiety on the A ring had little effect on anti-SOD activity. From these results, we conclude that substitution on the A ring has relatively little impact on the SAR.
1.1.6. The NMR Activity Inhibition by Low Molecular Weight Polyphenols
To determine what structural features of the flavonols contributed to their activities toward the enzyme, we investigated a variety of other low molecular weight compounds. Structures of compounds used in this study are presented in Figure 13. Of these compounds, only 4 and 22 (gallic acid and MTHB) showed significant inhibition at concentrations below 500 M. Compounds 3, 17 and 18 (IC50 = 3.0, 2.0, and 3.5 mM, respectively) were representative of those with the lower activities.
At pH 9 or below, 4 reacts with CuZnSOD in such a way as to prevent NMR relaxation activity of the enzyme. The reaction appears reversible, with an equilibrium constant of ~3 mM. At pH greater than 9, a second, irreversible reaction occurs. The rate of this second reaction is pH dependant in a way consistent with deprotonation occurring at the enzyme and not at the inhibitor.
1.1.7. Effects of 4 and 5 on SOD Activity at pH below ~9
We measured the effects of 4 concentration on the activity of CuZnSOD at pH ranging from 7.0 to 11.5. Our results indicate that a deprotonated form of the compound is responsible for SOD inhibition. At pH of 7 or less, 4 is predominantly charge neutral (pKa = 8.13) and a concentration of 4 mM does not appreciably diminish SOD activity. At pH 8 and 9, the activity of CuZnSOD decreased with increasing concentration of 4. A plot of the reciprocal of the normalized activity (A, defined in the experimental section) versus inhibitor concentration was linear (Figure 14), consistent with the reversible binding of the inhibitor to the enzyme. Results of a number of experiments indicate that NMR inhibition by 4 at pH 8 is reversible. For example, the fraction of enzyme inhibited by 4 decreased when samples were diluted, consistent (within experimental uncertainty) with the inhibition reversible ligand binding. In addition, removing MDHB by gel filtration restored relaxation activity of the enzyme. We note that these behaviors may arise due to addition of oxygen in the manipulation of samples.
Effects of pH on inhibition by 4: At pH greater than 10, a second reaction appears to occur between 4 and CuZnSOD. The kinetics of the second reaction could be conveniently monitored by NMR. Representative kinetic results are presented in Figure 15 for reaction of CuZnSOD with 1 mM 4. As shown in this figure, the data do not extrapolate back to 100% activity at the time of mixing (the zero point on the y axis). The activity of the enzyme at the intercept in Figure 15 is consistent with the equilibrium described above at pH 9.
At a constant 4 concentration (1.0 mM), the extrapolated value of the activity at the time of mixing was not
Figure 13. Structures of low molecular weight polyphenols used in this study.
Figure 14. At pH 8, MDHB (4) inhibits NMR relaxation of SOD in a concentration dependent manner. The trend line shows behavior for a reversible reaction, Kapp = 3.0 mM.
Figure 15. Normalized SOD activity decays with pseudo-first order kinetics in 1 mM4, pH 11.0.
influenced by pH over the range from 9 to 11.5 (data not shown) indicating that the binding constant is not grossly affected by pH. Continuation of the study to pH greater than 11.5 was not possible due to the pH dependent loss of enzyme activity documented elsewhere [62] . In contrast, the rate of the slower step increased dramatically with pH over this range (Figure 16). Since the pKa of 4 (reportedly 8.12, Perron, et al.) is well below the pH where the reaction becomes significant, the necessary deprotonation likely occurs at the enzyme. The trend line in Figure 12 shows the effect predicted if the pH dependence of the second step arose from a reaction requiring deprotonation of an SOD active site lysine with a reported pKa of 10.8 [62] .
When compared to 4, 5 is a much more effective inhibitor of SOD NMR relaxation activity at low concentration. The pronounced curvature of the plot suggests that 5 is strongly aggregated at mM concentration. If this is the case, then NMR inhibition at the lowest concentrations of 5 should best reflect the effects of the monomeric compound. The data from Figure 17 shows that inhibition by 5 was significant at micromolar concentrations of inhibitor. While gallols (such as 5) are better ligands than catechols (such as 4), the difference between the slopes of Figure 17 and Figure 18 at very low concentrations seems inconsistent with the effect arising due to differences in metal binding constants alone. Consistent with results of our EPR studies, we believe the difference in the redox behaviors of the two compounds accounts for the difference in their activities in the NMR assay: While 4 appears to inhibit the activity by binding to the enzyme, 5 effectively reduces the enzyme as well. Gallic acid (22) behaved similarly to 5.
1.1.8. Inhibitors of Zinc Metalloenzymes Were Ineffective against CuZnSOD
We examined the effects of a series of chelating agents that are known to inhibit zinc enzymes by binding the active site metal. While acetohydroxamic acid (25), maltol (26), thiomaltol (27), and 2-mercaptopyridine N- oxide (28) have been reported to inhibit zinc containing metalloproteinases with IC50 ranging from 25 mM to 35 M [63] , none significantly affected the NMR activity of CuZnSOD at mM concentrations.
Figure 16. Effect of pH on pseudo-first order rate constant (kobs) for the irreversible decomposition of the MDHB/CuZnSOD com- plex.
Figure 17. Effect of 5 concentration on SOD activity.
Figure 18. Chelating inhibitors of zinc containing metalloenzymes.
1.1.9. Inhibition of CuZnSOD NMR Activity by Catechol, 2- and 4-Mercaptophenol, and 1,2-Benzeneditiol
We measured the effects of 29, 30, and 31 (Figure 19) to investigate the effects of sulfur incorporation on the NMR inhibiting activity of catechol. The results of this experiment (presented in Figure 20), indicate an increase in potency with added sulfur. While this result could arise due to increased metal affinity [64] - [67] , it more likely reflects a shift in the redox potentials of the compounds.
Figure 19. Structures of 1, 2 disubstituted benzenes.
Figure 20. Anti-SOD activities of 29 (catechol), 30 (2-mercaptophenol) and 31 (1,2-benzenedithiol) increase with sulfur incorporation.
1.1.10. EPR Data for CuCl2 and CuZnSOD upon Addition of MTHB and MDHB
EPR data for CuCl2 and CuZnSOD upon addition of MTHB and MDHB. Numbers in brackets are estimated values.
asamples were prepared in Tris buffer (pH 8) or in glycine/NaOH buffer (pH 11) unless otherwise specified; bg value for MTHB or MDHB semiquinone radical; cestimated g and A values for Cu2+-buffer complex; destimated g and A values for MTHB, MDHB or EC hyperfine coupling with Cu2+.
NOTES
*Corresponding author.