Natural Science
Vol.5 No.6A(2013), Article ID:33032,5 pages DOI:10.4236/ns.2013.56A003
Mutations in FMN binding pocket diminish chromate reduction rates for Gh-ChrR isolated from Gluconacetobacter hansenii
![]()
1Fundamental & Computational Sciences Directorate, Pacific Northwest National Laboratory, Richland, USA; *Corresponding Author: hongjunj@mir.wustl.edu
2School of Public Health, University of Washington, Seattle, USA
3Department of Biochemistry, University of Texas Health Science Center, San Antonio, USA
4Washington University School of Medicine, Division of Radiological Sciences, St. Louis, USA
Copyright © 2013 Janin A. Khaleel et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received 16 April 2013; revised 18 May 2013; accepted 26 May 2013
Keywords: Bioremediation; Chromate Reduction; Enzyme Redesign; Site-Specific Mutagenesis; Chromate Reductase; Flavin Mononucleotide; Promiscuous Enzyme Activities
ABSTRACT
A putative chromate ion binding site was identified proximal to a rigidly bound FMN from electron densities in the crystal structure of the quinone reductase from Gluconacetobacter hansenii (Gh-ChrR) (3s2y.pdb). To clarify the location of the chromate binding site, and to understand the role of FMN in the NADPH-dependent reduction of chromate, we have expressed and purified four mutant enzymes involving the sitespecific substitution of individual side chains within the FMN binding pocket that form noncovalent bonds with the ribityl phosphate (i.e., S15A and R17A in loop 1 between β1 sheet and α1 helix) or the isoalloxanzine ring (E83A or Y84A in loop 4 between the β3 sheet and α4 helix). Mutations that selectively disrupt hydrogen bonds between either the N3 nitrogen on the isoalloxanzine ring (i.e., E83) or the ribitylphosphoate (i.e., S15) respectively result in 50% or 70% reductions in catalytic rates of chromate reduction. In comparison, mutations that disrupt π-π ring stacking interactions with the isoalloxanzine ring (i.e., Y84) or a salt bridge with the ribityl phosphate result in 87% and 97% inhibittion. In all cases there are minimal alterations in chromate binding affinities. Collectively, these results support the hypothesis that chromate binds proximal to FMN, and implicate a structural role for FMN positioning for optimal chromate reduction rates. As side chains proximal to the β3/α4 FMN binding loop 4 contribute to both NADH and metal ion binding, we propose a model in which structural changes around the FMN binding pocket couples to both chromate and NADH binding sites.
1. INTRODUCTION
Chromate [Cr(VI)] is a toxic pollutant that commonly leaches into ground water [1]. Recently it has been found that some bacteria express enzymes that are able to bind and reduce toxic Cr(VI) to form nontoxic Cr(III) as an intracellular precipitate, offering a cost-effective strategy of bioremediation [2,3]. Two enzyme classes are known to reduce chromate [3]. One class involves a metal reductase enzyme in the outer membrane that couples through a complex series of specialized electron transfer proteins [4-7]; the second class involves a cysosolic NAD(P)Hdependent chromate reductase (i.e., ChrR) that functions autonomously [2,8,9]. A major advantage of the latter class of enzymes is their ability to concentrate and sequester reduced chromate [i.e., Cr(III)] within the reducing environment of cells [10-12].
ChrR acts on quinones and other organic substrates (e.g., cancer prodrugs) through a ping-pong bi-bi (double displacement) mechanism whereby NAD(P)H and organic substrates bind sequentially, such that NAD(P)H reduces FMN prior to substrate binding and reduction [3,12-14]. In contrast, the reduction of chromate involves an ordered bireactant mechanism, whereby chromate must first bind prior to the association of NAD(P)H [8]. Following electron transfer and reduction of chromate, NAD(P)+ must dissociate prior to the release of the reduced chromate. Consistent with this latter mechanism, the crystal structure of ChrR (3s2y.pdb) suggests a likely ion binding site for chromate proximal to FMN within the NAD(P)H binding pocket [8]. As chromate reduction by ChrR involves an adventitious enzyme activity [9] with a low catalytic rate and binding affinity (kcat = 0.25 sec−1; Km = 0.24 mM) [8,13], the precise role of the FMN cofactor in chromate reduction remains uncertain. To test the prediction that chromate binds proximal to FMN, and to better understand the possible role of FMN in promoting the binding and reduction of chromate, we have used site-directed mutagenesis to assess how modifications in the FMN binding pocket affect chromate binding and its NADH-dependent reduction. Consistent with the proposal that FMN plays a catalytic role in chromate reduction, we observe substantial decreases in catalytic rates of chromate reduction upon the site-directed mutagenesis of four different FMN ligands (S15A, R17A, E83A, and Y84A) in the FMN binding pocket.
2. MATERIALS AND METHODS
2.1. Chemicals and Reagents
All chemicals were purchased from Sigma-Aldrich Fine Chemicals (St. Louis, MO) unless indicated otherwise. Ni-NTA resin was purchased from Qiagen Inc. (Valencia, CA). Isopropyl-D-1-thiogalactopyranoside (IPTG), agar, and LB medium were purchased from Fisher Scientific (Wilmington, MA). All plasmid purification kits were from Qiagen Inc. (Valencia, CA).
2.2. Site-Directed Mutagenesis
The DNA sequence of the Gh-ChrR gene from Gluconacetobacter hansenii ATCC 23769 (ZP_06834583), plus four single-codon mutants (S15A, R17A, E83A, Y84A and Y129N) were synthesized, and inserted into the expression vector pJexpress411 (DNA 2.0 Inc., Menlo Park, CA, USA) such that a 6-histidine tag was present at the C-terminus of the gene product. After that, mutant-expressing plasmids were codon optimized for expression in E. coli. All desired mutations were verified by DNA sequencing.
2.3. Protein Expression and Purification
Plasmids containing the gene encoding Gh-ChrR were transformed into E. coli BL21 (DE3) and recombinant protein expression was induced by the addition of IPTG to the medium (0.02 mM final concentration). Transformed BL21 cells in ice cold lysis buffer [0.3 M NaCl, 50 mM 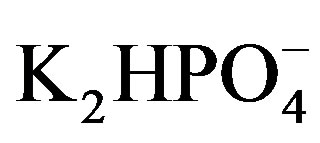 (pH 8.0)] were broken by repeated sonication (1 minute × 3) (Branson Ultrasonic, Danbury, CT). Supernatants were collected following centrifugation and protein was affinity purified using a Ni-NTA resin, essentially as previously described [8].
(pH 8.0)] were broken by repeated sonication (1 minute × 3) (Branson Ultrasonic, Danbury, CT). Supernatants were collected following centrifugation and protein was affinity purified using a Ni-NTA resin, essentially as previously described [8].
2.4. Chromate Reduction Activity Assays
Reduction activity of chromate by Gh-ChrR and its mutants were measured at 37˚C in 50 mM Tris-HCl (pH 7.4), 100 mM NaCl, 0.1 mM NADH, and 5 μM Gh-ChrR as a function of chromate concentration, where rates a NADH consumption were measured using absorbance changes at 340 nm, essentially as previously described [8]. Kinetic constants (Km and Vmax) were fit to a Michaelis-Menton equation. All measurements were conducted in triplicate.
3. RESULTS AND DISCUSSION
3.1. Inhibition of Chromate Reduction by Mutation of FMN Ligands
To investigate the role of FMN in the NADPH-dependent reduction of chromate, we have expressed GhChrR in E. coli and purified mutant enzymes involving site-specific substitution of side chains that respectively position FMN either through the ribityl phosphate (i.e., S15A and R17A in loop 1 between β1 sheet and α1 helix) or the isoalloxanzine ring (E83A or Y84A in loop 4 between the β3 sheet and α4 helix) (Figure 1). Following protein purifation, time-dependent rates of chromate [Cr(VI)] reduction were measured, permitting a determination of maximal catalytic rates (Vmax) and apparent affinities for chromate through measurements of the Michaelis-Menten constant (Km) (Figure 2). In all cases, mutant enzymes exhibited 50% or greater decreases in maximal catalytic rates of chromate reduction (Table 1).
3.2. Relationship between FMN Positioning and Chromate Binding
From the available crystal structure of ChrR (3s2y. pdb), the FMN cofactor is held within a pocket near the surface through noncovalent interactions within individual subunits involving 12 electrostatic and 5 hydrophobic contact interactions that primarily involve loops 1 (G14- SLRKASFN22) and 4 (P82EYNY86) [8]. The ribotyl phosphate group is positioned deep within the binding pocket, and is stabilized through a salt bridge with R17 in loop 1 between the β1 sheet and α1 helix (Figure 1), which is highly conserved in other NAD(P)H-dependent FMN reductases [13,15,16]. The proximal S15 within loop 1 appears to stabilize this interaction, acting to assist in positioning the ribotyl phosphate through the formation of a hydrogen bond. The stabilizing interactions between
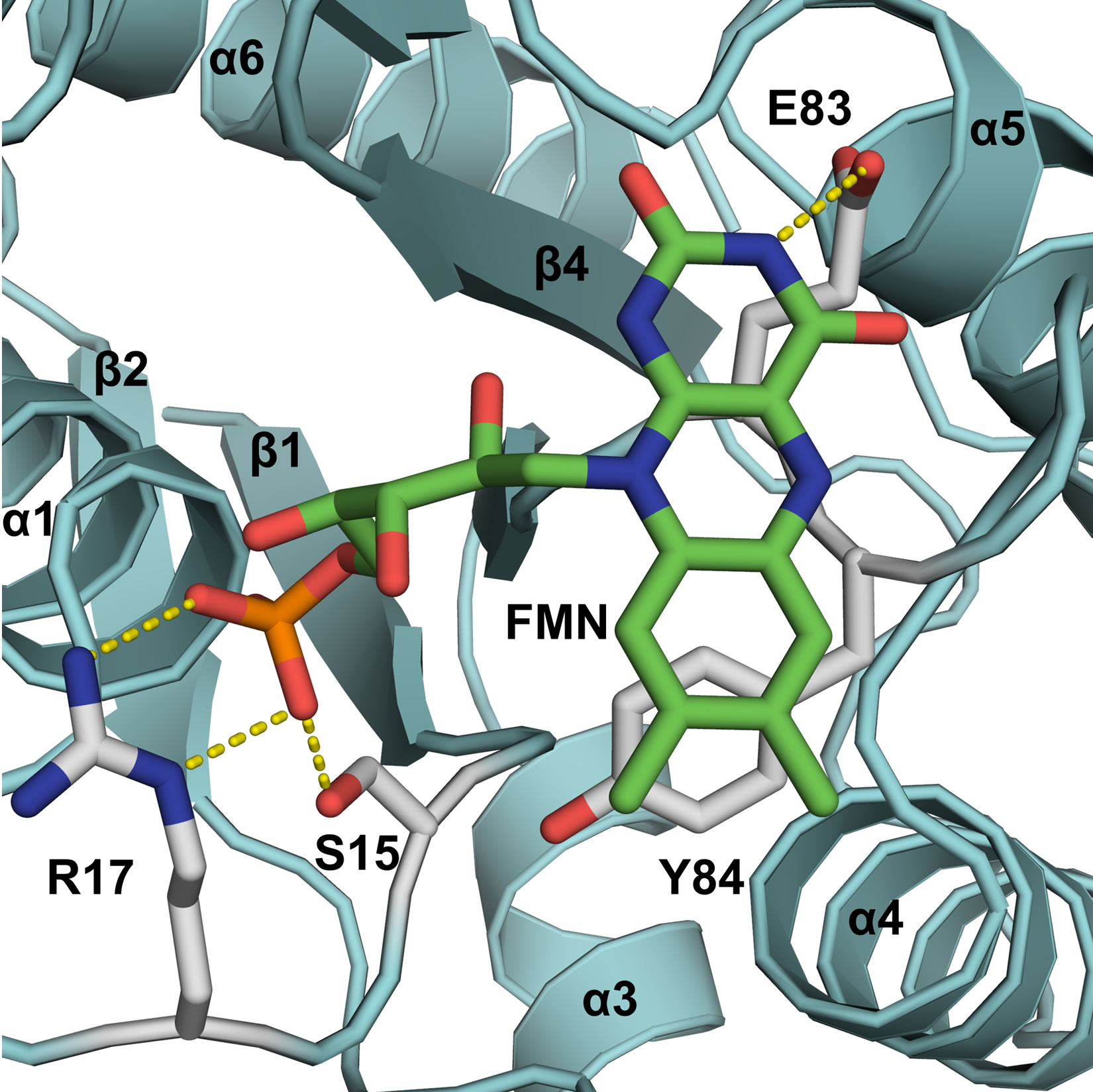
Figure 1. Mutant design for destabilization of the conformation of FMN within the binding cleft of GhChrR Monomers. FMN and side chain under investigation are depicted as sticks, highlighting electrostatic interactions (dashed yellow lines). Secondary structural elements of Gh-ChrR are shown schematically (blue). Site-specific mutations (underlined) were made in loop 1 (GSLRKASFN22) between the β1 sheet/α1 helix and loop 4 (PEYNY86) between the β3 sheet/α4 helix structural motifs, which respectively stabilize the ribityl phosphate or isoalloxanzine ring of FMN near the surface of individual subunits in the crystal structure of GhChrR (3s2y.pdb).
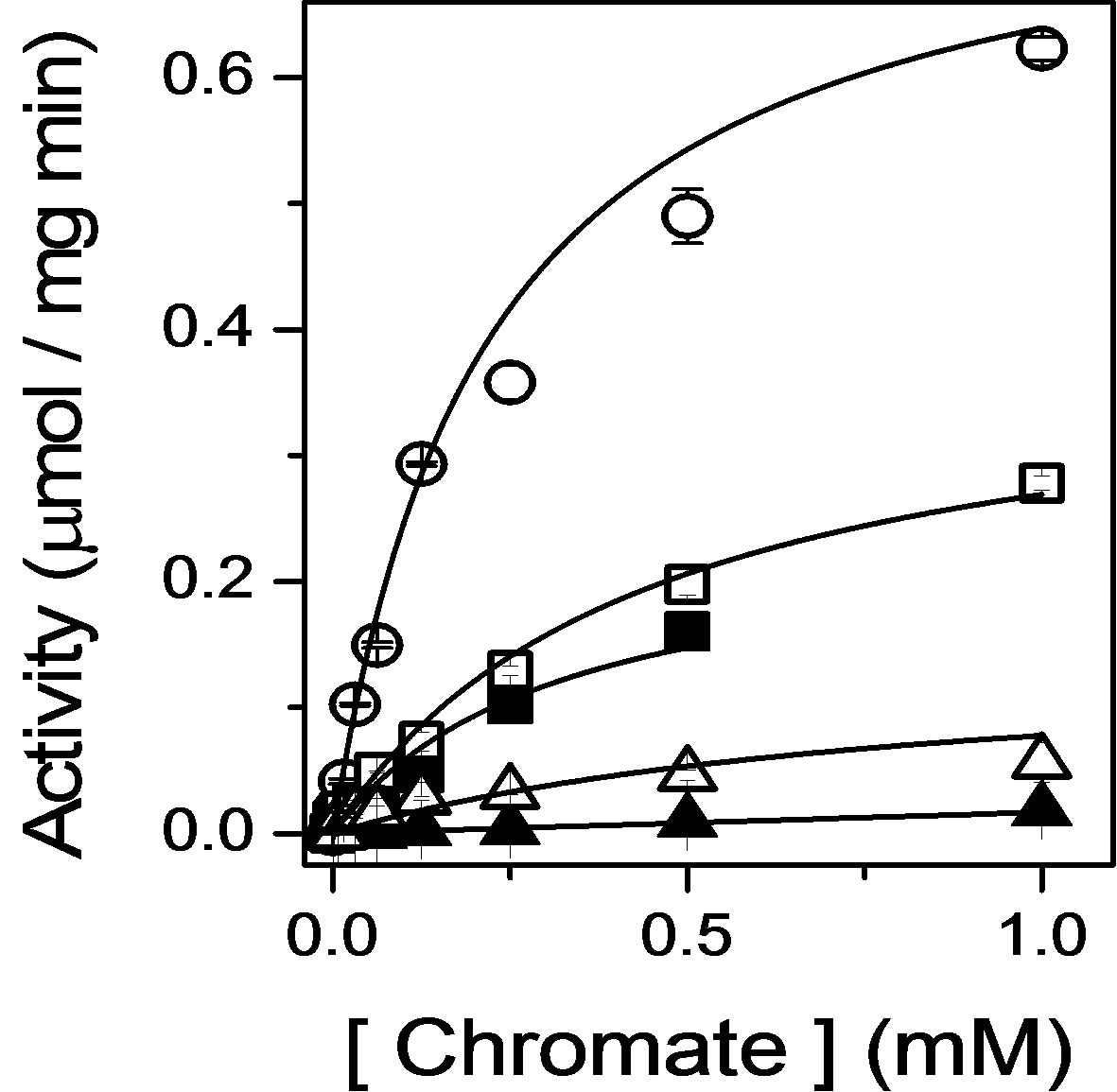
Figure 2. Inhibition of chromate reductase activity upon destabilization of bound FMN. Catalytic rates of reduction measured as a function of chromate, using absorbance changes of NADH, for wildtype Gh-ChrR (open circles), mutations in loop 1 (GSLRKASFN22) between the β1 sheet/α1 helix (S15A, closed squares; R17A, closed triangles), or mutations in loop 4 (PEYNY86) between the β3 sheet/ α4 helix structural motifs (E83A, open squares; Y84A, open triangles). Catalytic rates were measured at 37˚C in 50 mM Tris-HCl (pH 7.4), 100 mM NaCl, 0.1 mM NADH, and 5 μM Gh-ChrR.
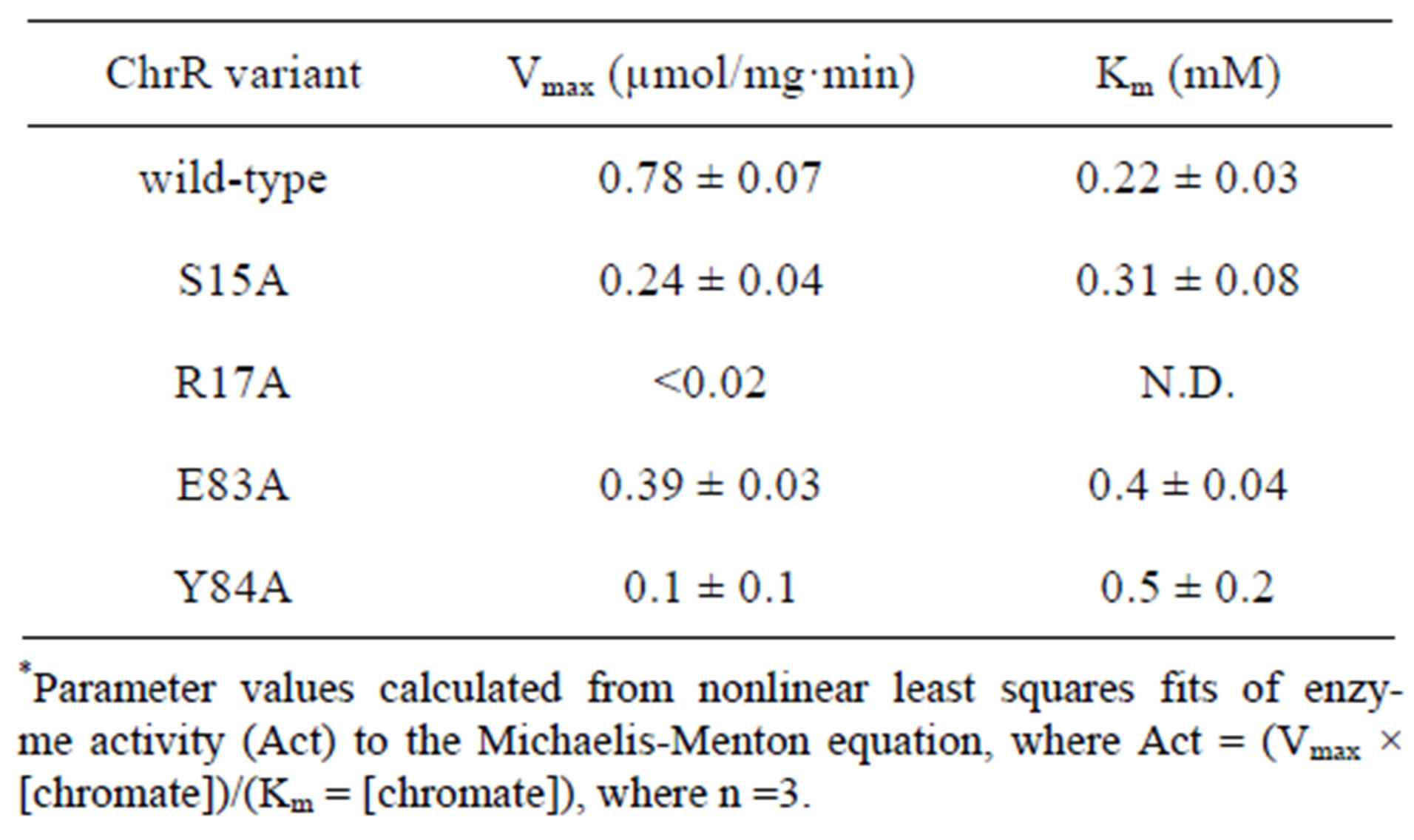
Table 1. Kinetic parameters for chromate reductase activity*.
loop 1 and the ribotyl phosphate are expected to be important for catalysis, as the isoalloxanzine ring of the FMN sits within a more hydrophobic cleft, whose positioning is likely dependent on the correct positioning of the ribotyl phosphate. Consistent with this expectation, mutations S15A and R17A respectively inhibit overall rates of chromate reduction by 70% and >97%.
Mutations E83A and Y84A respectively affect a hydrogen bond with the N3 nitrogen on the isoalloxanzine ring and a π-π stacking interaction that together positions the isoalloxanzine ring in the binding cleft, resulting in a 50% and 87% inhibition in the rates of chromate reduction. These latter results suggest that the orientation of the isoalloxanzine ring of FMN in the binding cleft plays an important role in modulating the overall rates of chromate reduction. This latter result is consistent with the putative assignment of the electron density proximal to the isoalloxanzine ring of FMN in the crystal structure as a metal binding site capable of binding chromate [8].
3.3. Proposed Structural Coupling between FMN Binding Cleft and Chromate Binding Site
As we have already described (see above), loop 4 (P82EYNY86) between the β3 sheet and α4 helix provides the ligands for the isoalloxanzine ring of FMN. This is significant, as loop 4 also contributes to NADH binding through the involvement of N85 [8] (Figure 3). Further, R101 in the α4 helix within this conserved FMN binding motif serves as a ligand to coordinate an anion (i.e., chloride) in the crystal structure of ChrR, and has been suggested to act as a primary ligand for chromate ion binding [8]. These latter suggestions are consistent with the experimental data, demonstrating similar decreases in catalytic rates upon mutations in either loop 1 or 4 that modify the positioning of the FMN cofactor, and suggest the presence of long-range structural effects through the FMN binding pocket that affect reduction at the chromate binding site. Further, such mutations may couple to modify NADH binding (not tested here), which may
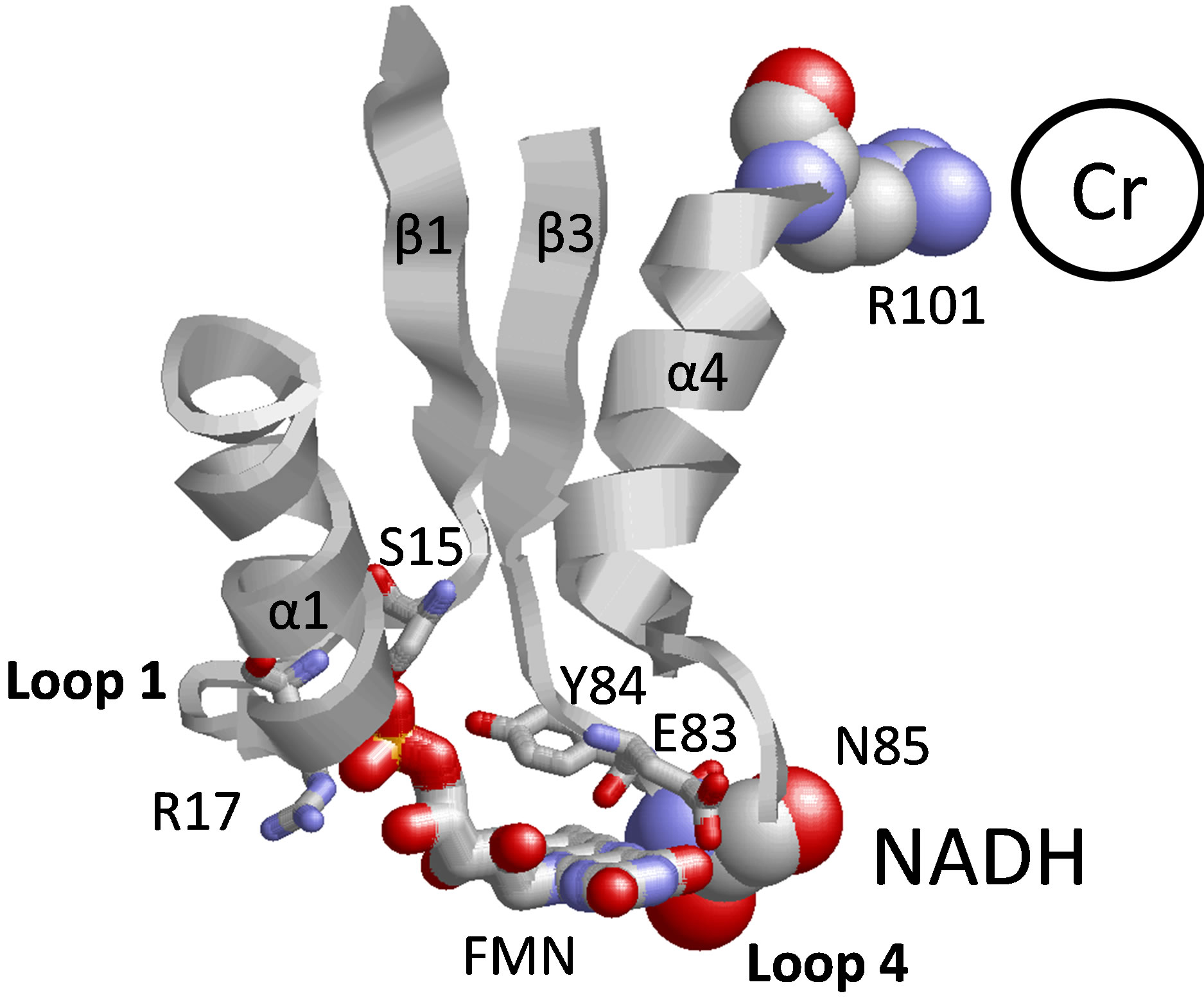
Figure 3. Proposed structural model between fmn binding cleft and amino acid side chains implicated in chromate reduction and NADH binding. Depicted are relative positions of FMN (wireframe image) in loop 1 (GSLRKASFN22) between the β1 sheet/α1 helix and loop 4 (PEYNY86) between the β3 sheet/α4 helix structural motifs (E83 and Y84 are shown). Mutated FMN ligand S15, R17 E83, and Y84 are shown in wireframe representation. Structural coupling between NADH and chromate binding is proposed through FMN binding cleft, involving NADH binding ligand N85 in loop 4 and chromate binding ligand R101 in α4 helix.
disrupt the NADH-dependent reduction of chromate. Such an allosteric coupling between the FMN binding cleft and NADH may represent part of a conformational switch that enables efficient NADH binding and subsequent release of NAD+ following the reduction of FMN during the normal turnover of ChrR in the presence of natural substrates (i.e., quinones); this mechanism may not apply in the NADH-dependent reduction of chromate. In this latter case, the ability of chromate to bind within this natural FMN binding cleft suggests design features that may further optimize chromate reductase activity involving, for example, site-specific mutations that enhance chromate binding while having minimal impact on FMN or NADH binding.
3.4. Conclusion and Future Directions
In summary, our results demonstrate substantial reductions in catalytic activity for four mutant enzymes involving ligands within the FMN binding cleft that selectively disrupt interactions with the ribotyl phosphate or isoalloxanzine ring, demonstrating an essential role for the positioning of the FMN cofactor for optimal rates of chromate reduction. The close positioning of NADH and the bound chromate suggests that the observed promiscuous chromate reduction activity does not directly involve the FMN cofactor. Rather, conserved structural elements within NAD(P)H-dependent FMN reductases may provide a structural framework for the construction of improved enzymes with catalytic activities appropriate for bioremediation (e.g., chromate reduction). Future measurements should design mutants that selectively increase chromate binding without significantly affecting the structures of either FMN or NADH binding clefts, and explore the possible direct electron transfer between NADH and bound chromate.
4. ACKNOWLEDGEMENTS
We thank David Kennedy and Andy Plymale for their technical support. This work was supported by Laboratory Directed Research and Development Funds (LDRD #90001) at Pacific Northwest National Laboratory (PNNL). Financial support comes principally from The US Department of Energy’s Office of Biological and Environmental Research. Battelle operates PNNL for the US Department of Energy under Contract DE-AC05-76RL01830.
REFERENCES
- Ackerley, D.F., Gonzalez, C.F., Keyhan, M., Blake II, R. and Matin, A. (2004) Mechanism of chromate reduction by the Escherichia coli protein, NfsA, and the role of different chromate reductases in minimizing oxidative stress during chromate reduction. Environmental Microbiology, 6, 851-860. doi:10.1111/j.1462-2920.2004.00639.x
- Ackerley, D.F., Gonzalez, C.F., Park, C.H., Blake II, R., Keyhan, M. and Matin, A. (2004) Chromate-reducing properties of soluble flavoproteins from Pseudomonas putida and Escherichia coli. Applied and Environmental Microbiology, 70, 873-882. doi:10.1128/AEM.70.2.873-882.2004
- Gonzalez, C.F., Ackerley, D.F., Lynch, S.V. and Matin, A. (2005) ChrR, a soluble quinone reductase of Pseudomonas putida that defends against H2O2. Journal of Biological Chemistry, 280, 22590-22595. doi:10.1074/jbc.M501654200
- Xiong, Y., Chen, B., Shi, L., Fredrickson, J.K., Bigelow, D.J. and Squier, T.C. (2011) Targeted protein degradation of outer membrane decaheme cytochrome MtrC metal reductase in Shewanella oneidensis MR-1 measured using biarsenical probe CrAsH-EDT(2). Biochemistry, 50, 9738- 9751. doi:10.1021/bi200602f
- Shi, L., Squier, T.C., Zachara, J.M. and Fredrickson, J.K. (2007) Respiration of metal (hydr)oxides by Shewanella and Geobacter: A key role for multihaem c-type cytochromes. Molecular Microbiology, 65, 12-20. doi:10.1111/j.1365-2958.2007.05783.x
- Xiong, Y., Shi, L., Chen, B., Mayer, M.U., Lower, B.H., Londer, Y., Bose, S., Hochella, M.F., Fredrickson, J.K. and Squier, T.C. (2006) High-affinity binding and direct electron transfer to solid metals by the Shewanella oneidensis MR-1 outer membrane c-type cytochrome OmcA, Journal of the American Chemical Society, 128, 13978- 13979. doi:10.1021/ja063526d
- Shi, L., Chen, B., Wang, Z., Elias, D.A., Mayer, M.U., Gorby, Y.A., Ni, S., Lower, B.H., Kennedy, D.W., Wunschel, D.S., Mottaz, H.M., Marshall, M.J., Hill, E.A., Beliaev, A.S., Zachara, J.M., Fredrickson, J.K. and Squier, T.C. (2006) Isolation of a high-affinity functional protein complex between OmcA and MtrC: Two outer membrane decaheme c-type cytochromes of Shewanella oneidensis MR-1. Journal of Bacteriology, 188, 4705-4714. doi:10.1128/JB.01966-05
- Jin, H., Zhang, Y., Buchko, G.W., Varnum, S.M., Robin son, H., Squier, T.C. and Long, P.E. (2012) Structure determination and functional analysis of a chromate reducetase from Gluconacetobacter hansenii. PLoS One, 7, Article ID: e42432. doi:10.1371/journal.pone.0042432
- Gonzalez, C.F., Ackerley, D.F., Park, C.H. and Matin, A. (2003) A soluble flavoprotein contributes to chromate reduction and tolerance by Pseudomonas putida. Acta Biotechnologica, 23, 233-239. doi:10.1002/abio.200390030
- Park, C.H., Keyhan, M., Wielinga, B., Fendorf, S. and Matin, A. (2000) Purification to homogeneity and characterization of a novel Pseudomonas putida chromate reductase. Applied and Environmental Microbiology, 66, 1788-1795. doi:10.1128/AEM.66.5.1788-1795.2000
- Barak, Y., Ackerley, D.F., Dodge, C.J., Banwari, L., Alex, C., Francis, A.J. and Matin, A. (2006) Analysis of novel soluble chromate and uranyl reductases and generation of an improved enzyme by directed evolution. Applied and Environmental Microbiology, 72, 7074-7082. doi:10.1128/AEM.01334-06
- Barak, Y., Thorne, S.H., Ackerley, D.F., Lynch, S.V., Contag, C.H. and Matin, A. (2006) New enzyme for reductive cancer chemotherapy, YieF, and its improvement by directed evolution. Molecular Cancer Therapeutics, 5, 97- 103. doi:10.1158/1535-7163.MCT-05-0365
- Eswaramoorthy, S., Poulain, S., Hienerwadel, R., Bremond, N., Sylvester, M.D., Zhang, Y.B., Berthomieu, C., Van Der Lelie, D. and Matin, A. (2012) Crystal structure of ChrR—A quinone reductase with the capacity to reduce chromate. PLoS One, 7, Article ID: e36017. doi:10.1371/journal.pone.0036017
- Thorne, S.H., Barak, Y., Liang, W., Bachmann, M.H., Rao, J., Contag, C.H. and Matin, A. (2009) CNOB/ChrR6, a new prodrug enzyme cancer chemotherapy. Molecular Cancer Therapeutics, 8, 333-341. doi:10.1158/1535-7163.MCT-08-0707
- Liger, D., Graille, M., Zhou, C.Z., Leulliot, N., Quevillon-Cheruel, S., Blondeau, K., Janin, J. and van Tilbeurgh, H. (2004) Crystal structure and functional characterization of yeast YLR011wp, an enzyme with NAD(P)HFMN and ferric iron reductase activities. Journal of Biological Chemistry, 279, 34890-34897. doi:10.1074/jbc.M405404200
- Drennan, C.L., Pattridge, K.A., Weber, C.H., Metzger, A.L., Hoover, D.M. and Ludwig, M.L. (1999) Refined structures of oxidized flavodoxin from Anacystis nidulans. Journal of Molecular Biology, 294, 711-724. doi:10.1006/jmbi.1999.3151

