1. Introduction
Species from Aspergillus section Flavi have the potential to contaminate agricultural commodities with carcinogenic mycotoxins such as aflatoxins. Research is ongoing to understand and prevent aflatoxin contamination, and thereby ensure the safety of our food and feed supply. Nearly a decade has passed since two pre-harvest biopesticides (AF36 and Afla-Guard®) were licensed by the Environmental Protection Agency (EPA) for use as biocontrol agents to prevent pre-harvest aflatoxin contamination. AF36 and the component strain in Afla-Guard® are A. flavus isolates that are unable to produce aflatoxins (AF−) because of either mutations in, or lack of, critical genes required for aflatoxin biosynthesis. For AF36, a point mutation in the aflatoxin biosynthesis gene pksA (aflC) is responsible for the AF− phenotype [1], while the Afla-Guard® strain (NRRL21882) is missing most of the genes in the 80 kb aflatoxin biosynthesis gene cluster up to the proximal telomere on chromosome 3 [2,3]. Field trials have shown that proper application of spores from these fungi greatly reduces the incidence of aflatoxin contamination in corn [4], cottonseed [5], and peanuts [6], and is being considered for use against contamination of tree nuts. The ability of these strains to reduce aflatoxin contamination has been postulated to be due to displacement of the aflatoxin-producing populations in the soil [5]. An alternate explanation is that contact (thigmotropism) between the AF− isolate and an aflatoxigenic (AF+) isolate causes the inhibition [7]. Other candidate strains for biocontrol are being developed for use on corn (K49 in Mississippi and KD17, KD19, and KD22 in Louisiana and Texas). All of these A. flavus strains produce large sclerotia (L-strain), and contain the Mat1-2 mating type idiomorph.
AF36 and K49 share identical sequence with regard to the pksA polymorphism, but K49 possesses a truncated hybrid polyketide synthase-nonribosomal peptide synthase gene in its cyclopiazonic acid (CPA) gene cluster, resulting from a chance mutation that introduces a stop codon [8], that renders it unable to synthesize CPA. Although AF36 retains the ability to produce CPA it is used for prevention of AF contamination in cotton, figs, and pistachios in Arizona and California [9,10]. The application substrate during treatment involves sterilized wheat seeds that have been colonized by the AF36 strain, and 10 pounds per acre is applied annually with an estimated cost of between $6 and $15 per acre [9]. The component strain in Afla-Guard®, NRRL21882, was first isolated from a peanut field in Georgia [11] and was initially used to protect peanut crops in Georgia. Since 2010, it has been marketed by Syngenta as a biocontrol strain for corn [12]. The application substrate involves hulled barley, rather than wheat, that has been colonized by NRRL21882. The annual application rate is 20 pounds, with an estimated cost of between $16 - $30, per acre [9]. Though not yet commercially available, K49 shows promise as a candidate strain for biocontrol of aflatoxin contamination of corn [12]. Biodegradable plastic has been used as an inexpensive substrate for production of the inoculum [13], but the application rate and the estimated cost for a grower to apply K49 each year has not yet been determined. KD17, KD19, and KD22 were isolated from a corn kernels harvested in Louisiana and Texas. The genetic basis for the AF− phenotype of these strains is uncertain. At this time, laboratory and greenhouse tests found that combined application of either KD17 or KD19 with NRRL21882 offered a more robust prevention of aflatoxin contamination compared to that achieved by application of each strain individually (Kenneth Damann, personal communication).
The long-term impact of introducing large amounts of these strains into fields to compete with native field populations of fungi has not been carefully studied. If the strains displace the native aflatoxin-producing population then there should not be a need to treat the fields annually, which is not the current practice. This consideration is of particular concern for control of aflatoxin contamination of corn where contamination usually does not occur every year. Another important consideration is that recombination may occur in natural populations of A. flavus [3]. There is in vitro evidence that mating can occur between individuals from natural field populations of Aspergillus, both within [14-16] and between [17] aflatoxin-producing species; furthermore, aflatoxigenicity can be re-introduced into the biocontrol strain by recombination [18]. This consideration is important because there is a risk that a recombinant AF+ strain may have an even greater potential for contamination as a result of its unique ability to out-compete native populations of fungi. Annual applications of the biocontrol strain may partly offset such a possible scenario, but this approach may become ineffective after multiple years of reapplication.
To determine the longevity of biocontrol populations introduced into the field and their ability to out-cross after application, we have developed fluorescent-tagged biocontrol strains of the most commonly used isolates. Our current study explores the ability of such biocontrol strains to be recovered after exposure to non-fluorescent biocontrol strains or wild-type aflatoxin-producing strains when the competition is performed under controlled laboratory conditions. This study is meant to be preliminary to assessing recovery of similar strains introduced into agricultural fields, and to tracking the ability and frequency of out-crossing of these strains with natural populations. Based on this study we find that introduction of a fluorescent marker into most of these biocontrol strains does not impair their abilities to self-compete or to compete with aflatoxin-producing strains.
2. Materials & Methods
2.1. Biocontrol and Competitor Strains
Strain NRRL21882 was obtained from the SRRC fungal collection. The AF36 strain was purchased from the ARS Culture Collection at the National Center for Agricultural Utilization Research (NCAUR) in Peoria, IL. The K49 and three KD strains were acquired from Dr. Kenneth Damann at Louisiana State University. For all strains, the species designation as A. flavus species that are considered large-sclerotium producers, as well as their mating-type idiomorph, had been previously reported or examined. The toxigenic strains used for the competition experiment included two A. flavus strains (SRRC1000-F, SRRC41) and three A. parasiticus strains (SRRC143-A, SRRC1032, SRRC2043). Information for the toxin profiles of all strains is shown in Table 1.
2.2. Molecular Investigation
For each wild type isolate, a flask containing 75 mL of potato dextrose broth was inoculated with 500 µl of fungal spore suspension. Flasks were agitated in an orbital shaker at 30˚C (130 rpm) for approximately 24 hours to accelerate the growth of mycelia. DNA was then extracted from harvested mycelia using the MasterPureTM Yeast Purification Kit (Epicentre Biotechnologies). The main genomic region of interest for this study is an upstream portion of the aflatoxin cluster between the aflF (norB) and aflU (cypA) genes. It has been reported that not only can the aflF/aflU region be used to segregate B and G aflatoxin producers [19] it can also offer information relating to sclerotial genotype for A. flavus [20]. To confirm the genetic basis for the sclerotial genotype of each strain, the intergenic region between the aflatoxin cluster genes aflF and aflU was amplified using the protocol of Ehrlich and co-workers [19]. An A. flavus L-strain genotype will result in an amplicon size of ap-
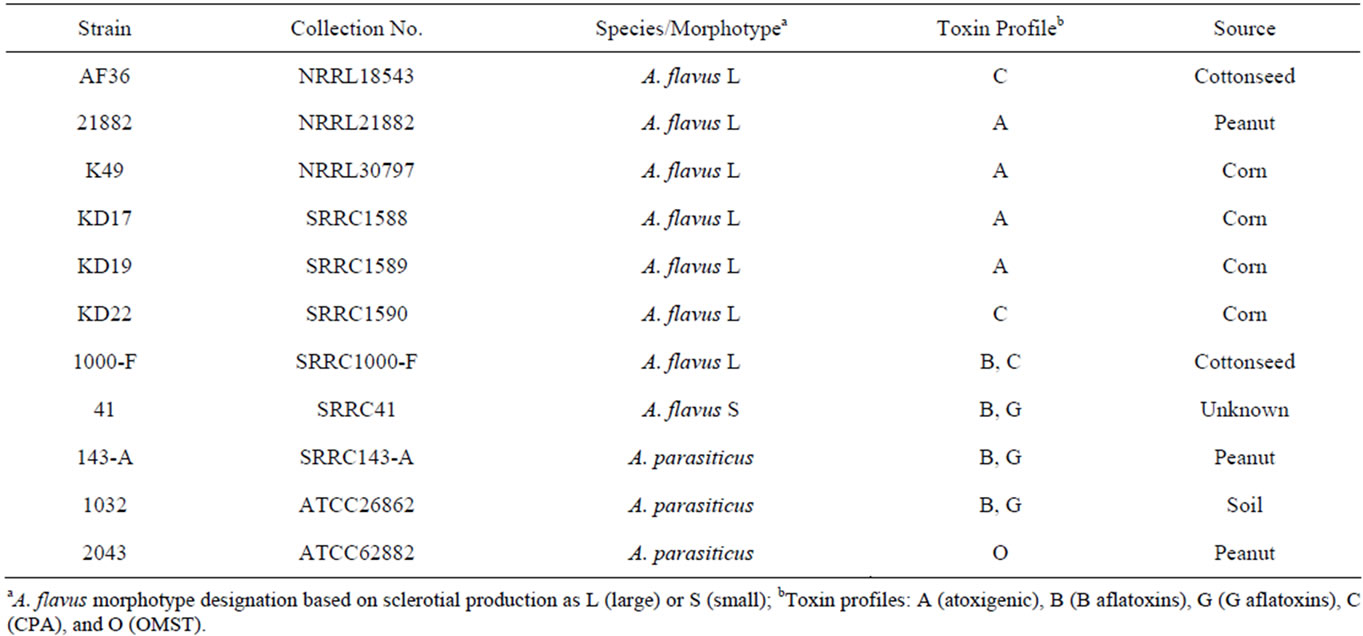
Table 1. Species and toxin information for competitor strains.
proximately 1 Kb, whereas an SB-strain (producer of only B aflatoxins) genotype will amplify a product of approximately 300 bp. Any isolate that produces B and G aflatoxins (A. parasiticus or A. flavus SBG) will result in an amplicon size of approximately 1.8 Kb. The mating type idiomorph for each strain in this study was also investigated following the diagnostic protocol as in Ramirez-Prado et al. [21].
2.3. Fungal Transformation with eGFP
The six AF− strains were co-transformed with pPTRI (Takara) and puc18-A.n.-GPD promoter-eGFP-nmt1 terminator [22] and selected for on pyrithiamine media. Transformations were done as previously described [23]. Fluorescent transformants were single-spored twice.
2.4. Experimental Design
A preliminary comparison for stability of the wild type (W) and transformed (T) isolates was performed for the biocontrol strains AF36 and NRRL21882 involving the following pairs: AF36 T/AF36 W, NRRL21882 T/NRRL21882 W, AF36 T/NRRL21882 W, and NRRL21882 T/AF36 W. A total of 104 spores per mL, from each pair of fungi, were co-inoculated on a potato dextrose agar plate and incubated in darkness at 30˚C. Resulting conidia were sampled from the colonies at 2, 6, 13, and 20 days after inoculation, and aliquots were diluted in 0.01% Triton X-100 for re-inoculation of colonies, developing from single spores, on Czapek’s (CZ) agar. After two days of incubation at 30˚C, the resulting colonies were examined for fluorescence and then counted. The next experimental component was to determine whether or not the eGFP transformation would 1) be successful and fluoresce, and 2) affect the individual growth of the six AF− strains. CZ plates were singlepoint inoculated for each W individual as well as each T individual. The plates were incubated in darkness at 25˚C for approximately eight days. Starting at day 4, each of the colonies was measured, to compare the W and T growth rates, and photographed in both white light and long-wave UV light (365 nm) to compare fluorescence. The final component of this study sought to test the aggressiveness of each T strain in the presence of host tissue and a competing microbe such as a its W parent, a different T strain or a toxigenic strain. This involved a single, surface sterilized (40% bleach with 0.001% Triton), and delinted cottonseed (Stoneville 7A) embedded in the center of a CZ plate. The competitor strains were then equidistantly single-point inoculated on opposite sides of the cottonseed with standard spore concentrations ranging between 1.35 and 2.6 × 106 spores per mL. Homologous comparisons involved each T strain competing against its W parent (e.g. AF36 T vs AF36 W), and heterologous comparisons involved each T strain competing against all other strains (e.g. AF36 T vs 1000-F). Assuming the growth rate for each AF− strain (T and W) would be similar, only the T AF− strains were included in the heterologous comparisons. Five replicate plates (labeled d4 - d8) for each competition experiment were incubated in darkness at 25˚C. Starting at day 4, all d4 plates were measured and photographed as mentioned above. Each subsequent day an additional replicate plate was examined (day 5 = d4 + d5 plates, etc.) until day 8 when all five replicate plates were measured and photographed. When either strain was the first to reach the cottonseed, the measurements for that plate would cease. If after day 8 neither strain reached the cottonseed, the measurements and photographs continued to a maximum of 11 days. If at any point a contaminant interfered with either competing colony or colonized the cottonseed before either competitor, that plate was removed and fewer replicate plates were used for further analyses. Once measurements for all non-contaminated plates were complete, the areas for each competitor colony (all replicates) were averaged and an average percent difference (APD) was calculated to ascertain which strain exhibited more robust growth in the presence of host tissue. To calculate APD, the averaged colony areas are subtracted from one another; the difference is then divided by the sum of the two averaged colony areas and multiplied by 100. A statistical two-tail T-test was then used to determine the significance of the difference in colony areas for each of the two competing strains.
3. Results
3.1. Molecular Investigation
Six atoxigenic A. flavus strains, currently in use for biocontrol in several laboratories, were compared in this study (AF36, NRRL21882, K49, KD17, KD19 and KD22). Previous examination of the aflF/aflU region in AF36 showed it to have a 1.5 kb deletion typical of A. flavus SB strains (the SB sequence motif) [20] rather than the smaller sequence deletion (1 kb) typical of isolates with L-strain sclerotia. This was confirmed in our investtigation. Two other AF− strains (K49 and KD19) exhibit the SB sequence deletion while KD22 exhibits the Lstrain deletion. KD17 exhibited no amplicon band upon PCR with primers to the aflF/aflU region and like strain NRRL21882 may lack this portion of the AF biosynthesis gene cluster. The toxigenic A. flavus, isolate SRRC1000-F, used in this study exhibits an L-strain genotype, and isolate SRRC41 exhibits an SBG phenotype with an intact aflF/aflU region (1.8 kb) like that of A. parasiticus. All but two strains in this study (SRRC41 and A. parasiticus SRRC143-A) were determined to have the Mat1-2 mating type idiomorph. These results, as well as the results for the mating type diagnostics of these strains, are shown in Table 2.
3.2. Preliminary Comparisons for Transformed AF36 and NRRL 21882
The method used for preliminary comparison of the fluorescent transformants and the wild-type parents involved mixing spores of W and T fungi (AF36 and NRRL21882) on a CZ plate and examining them for recovery after outgrowth for up to three weeks. Neither co-inoculated AF36 strain (W or T) showed a significant

Table 2. Sclerotial genotype and MAT idiomorph for competitor strains.
difference (Figure 1) in recovery over time, whereas NRRL21882 T was recovered in significantly reduced yield over time when co-inoculated with NRRL21882 W and with AF36 W. Similar comparison of AF36 T and NRRL21882 W showed that by day 10 recovery from AF36 colonies was almost three-fold greater than that of NRRL21882.
3.3. Individual Growth for Transformed AF− Strains
Figure 2 shows the colony morphology for each W and T AF− strain at day 4 (A group), and again at day 8 (B group), on CZ medium. At day 4, both AF36 and K49 appear to have similar morphology and growth rate (A1 and A3, respectively). The W colonies appear round, mostly vegetative (white), with little conidiation. The T colonies, though round, are somewhat smaller in diameter (indicating a slight reduction in growth rate) than the W colonies, but the colonies appear to exhibit slightly enhanced conidiation. The fluorescence for both T strains was easily observed and strong. At day 8 the W colonies were still round, and slightly larger than the T colonies, but subtle differences began to appear in the degree of conidial pigmentation (B1 and B3). There is a ring of conidia that appears darker and more defined for the A36 colonies (B1a, b). Though still highly fluorescent, AF36 T shows a lower observable fluorescence in the colonies with darker conidia both at the point of inoculation as well as in a ring around the point of inoculation (B1c)