Molecular Detection of Resistance to Rifampin and Isoniazid among Patients Eligible for Retreatment Regimen in Côte d’Ivoire in 2012 ()
1. Introduction
Drug-resistant tuberculosis has emerged as an important global public health threat. Rapid diagnosis of tuberculosis and multidrug-resistant (resistance to at least Rifampin and Isoniazid) is an important challenge to ensure a rapid and adequate therapy of tuberculosis and to limit the dissemination of multiresistant strains [1]. Rifampin and Isoniazid are the cornerstones of first-line drugs used for treatment of tuberculosis. In poor countries where TB is endemic, bacteriological confirmation of tuberculosis is carried out in most cases after Ziehl-Neelsen staining. Culture and drug-susceptibility testing are not available consequently the essential information for detection, follow-up and management of antibiotic resistance in M. tuberculosis cannot provide by laboratories.
Prevalence of MDR-TB has been estimated to be low in sub-Saharan Africa, where surveillance of drug resistance is limited notably [1]. In Côte d’Ivoire, prevalence of MDR-TB cases among new cases with a positive smear decreased from 5.3% to 2.5% respectively from 1995 to 2006 [2].
For patients previously treated for tuberculosis, there are few data notably for the principal two first-line drugs in Côte d’Ivoire contrary to new tuberculosis cases with positive smears. It has been shown that patients with a history of previous tuberculosis therapy have more risk to develop tuberculosis with resistant strains [3,4].
New diagnostic tools for tuberculosis were developed and validated, particularly molecular methods [5-8]. Molecular methods are highly efficient for Rifampin resistance detection and well adapted to search for the most relevant Isoniazid resistance. Some methods require minimal infrastructure and equipments [9].
Study objective was to describe the susceptibility profile to Rifampin and Isoniazid among previously treated patients who are eligible for retreatment and who had received the 2RHZES/1RHZE/5HRE regimen in Côte d’Ivoire with a molecular method in 2012.
2. Methods
2.1. Study Sites
Patients were recruited in reference regional Centre for tuberculosis in Côte d’Ivoire which represents intermediate level of sanitary pyramid. Symptomatic patients were screened based on their history [10,11]. Data collected included age, gender, previous anti-TB treatment and results of their smears for AFB detection after ZiehlNeelsen colouring.
Sputum decontamination and molecular assay were performed at Reference National Tuberculosis Laboratory located at Institut Pasteur de Côte d’Ivoire (Abidjan) which is renovated and equipped.
2.2. Samples Collection and Transport
For each patient enrolled, two sputum samples (spot, early morning) were collected and put in individual bag. In District of Abidjan sputum samples were transported daily to the laboratory refrigerated at +4˚ before transported in cool boxes to the Mycobacteria Laboratory. For the other region sputum samples were conserved at −20˚ and transported once per month to tuberculosis laboratory.
2.3. Resistance Detection
2.3.1. DNA Extraction
Sputum samples were processed using the U.S. Centers for Disease Control and Prevention (US CDC) recommended method of N-acetyl-L-Cystein 4% NaOH-2.9% Citrate. Five milliters of sputum were transferred in 50 ml of plastic centrifuge tube. Five milliters of NALC solution were added to sputum. Preparation was mixed by vortexing for 1minute and incubated at room temperature for 15 minutes. In the centrifuge tube, 35 ml of sterile phosphate buffer pH 6.8 were added and centrifuged at 3000 × g for 20 minutes. Supernatant was carefully eliminated. Pellet was resuspended with 2 ml of sterile phosphate buffer. Two hundred microliters of resuspended pellet were used to perform a smear which was stained by Ziehl-Neelsen method. Five hundred microliters of sediment was transferred in eppendorf tube of 1500 µl. Suspension was centrifuged at 10,000 × g in aerosol-tight rotor during 15 minutes. Supernatant is discarded. One hundred microliters of molecular water was added to preparation. Bacterial pellet was resuspended by vortexing. Bacterial suspension was inactivated at 95˚C and incubated in ultrasonic bath respectively for 20 minutes and 15 minutes. Bacterial suspension inactivated was centrifuged at 13,000 × g for 5 minutes. Supernatant containing DNA was transferred into fresh tube.
For DNA extraction, a negative control was included in each run of sputum sample decontaminated.
2.3.2. Amplification of DNA Extracted from Sputum Samples
With sputum containing AFB, Genotype MTBDR assay version 1.0 (Hain Lifescience, Nehren, Germany) was performed as recommended by the manufacturer. The amplification mixture contained 35 µl of primer-nucleotide mix provided in the kit, 5 µl of 10 × Taq polymerase incubation buffer containing 2 mM of MgCl2, 1 to 2 unit(s) of thermostable Taq DNA polymerase, and 5 µl of extracted chromosomal DNA solution in a final volume of 50 µl. Amplification parameters used were: 5 min of denaturation at 95˚C, followed by 10 cycles of 30 s at 95˚C and 2 min at 58˚C, followed by 30 additional cycles of 25 s at 95˚C, 40 s at 53˚C, and 40 s at 70˚C, ending with a final extension step of 8 min at 70˚C.
Prior to hybridization, a migration of 5 µl of each PCR products was performed during 30 minutes at 100 volts in 1% agarose gel stained with ethidium bromide. DNA amplified was revealed with a UV Transilluminator Lamp (365 nm).
2.3.3. Hybridization
Hybridization and detection were performed with a Twin Cubator semiautomated washing and shaking device according to the manufacturer’s instructions and using the reagents provided with the kit. Twenty microliters of denaturation solution was mixed to 20 µl of amplified sample. Mixed solution was incubated at room temperature for 5 min. One milliliter of prewarmed hybridization buffer was added before the membrane strips were placed and shaken in the hybridization solution for 30 min at 45˚C. After two washing steps, a colorimetric detection of the hybridized amplicons was obtained by the addition of the streptavidin alkaline phosphatase conjugate.
An internal quality control program with positive and negative controls was implemented during the study. An interpretable MTBDRplus assay was defined as a test strip with all control markers positive, including results of the markers for positive control (H37Rv strain), negative control for DNA extraction and for mix preparation.
2.4. Statistical Tests
Data were entered in MS Excel 8.0 and analyzed using Epi-info 6.04 (CDC, Atlanta). The Rate of Resistance to Rifampin and Isoniazid in the category of the patients recruited with his 95% confidence interval (CI) were estimated. The rates obtained were compared using Chisquare testing.
3. Results
For each run of DNA extracted, analysis of the PCR products with a UV Transilluminator Lamp revealed no presence of DNA for each negative control used (Figure 1).
A total of 278 patients were enrolled into the study. These previously treated patients were all pulmonary tuberculosis cases with positive smear for AFB. AFB detection on concentrated sputum showed that majority of samples analyzed were interpreted at least 2+. Sputum with exact number for 100 fields was found in recurrent TB cases (Table 1). Among them, there was 148 (53.2%) recurrent TB cases, 118 (42.5%) failures cases, and 12 (4.3%) defaulted from treatment.
Repartition of included patients according to the gender showed that 190 (68.3%) were males and 88 (31.7%) females. The majority of previously treated patients were in the 25 - 34 age-group (Table 2).
Among sputum samples collected from previously treated patients, a monoresistance to Rifampin and Isoniazid were confirmed respectively for 20 and 9. For the infectious strains excreted by patients, 101 (36.3%) were declared susceptible to Rifampin and Isoniazid.
From sputum of previously treated patients, the resistance to Isoniazid associated to resistance to Rifampin was diagnosed for 148 patients (53.2%, 95% CI, 47% - 59%) of them (Table 3).
In sputum of patients treated with the 2RHZE/4RH regimen, among the failures cases, resistance to Rifampin and Isoniazid was observed in 60 (69.8%, 95% IC, 60% - 80%).
For failures cases to the 2RHZES/1RHZE/5HRE regimen, among 32 sputum samples received 24 (75%, 95% IC, 60% - 90%) were declared resistance to Rifampin and Isoniazid.
Resistance to Rifampin and Isoniazid was detected in 60 (41%, 95% IC, 33% - 49%) sputum samples of 148 recurrent TB cases. Among the 12 cases of defaulted from treatment, 4 (33.3%) sputum samples contained strains resistant to Rifampin and Isoniazid.
There was no statistically significant difference for rates of Rifampin resistance detected from sputum samples in the various cases of failures with 2RHZE/4RH and RHZES/1RHZE/5HRE regimens. The comparison of resistance rates to Rifampin estimated from sputum samples in the 2 cases of treatment failures and the recurrence tuberculosis cases showed a statistically significant difference (Table 4).
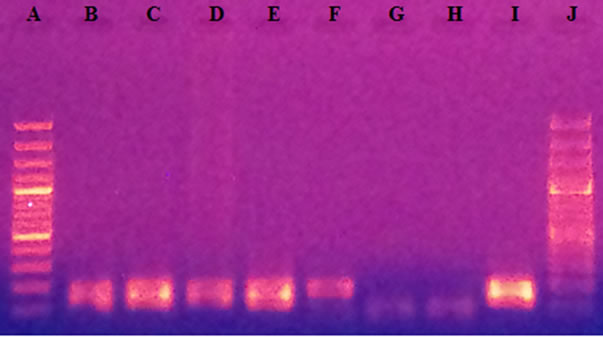
Lines A and J: Molecular weight Marker (100pb); Lines B, C, D, E, F: amplified DNA of sputum samples; Line G: Negative control of mix preparation; Line H: Negative control of DNA extraction; Line I: amplified DNA of H37rv.
Figure 1. Revelation of PCR products after migration.

Table 1. Results of microscopic examination according to the scale of WHO and IUATLD [12].
4. Discussion
In this study, an in vitro method for amplifying target DNA sequences was used for the diagnostic of tuberculosis and drug susceptibility testing. For the method chosen, before extraction of DNA, sputum specimens containing live bacilli and normal flora must be decontaminated using the N-acetyl-L-cystein (NALC)-sodium hydroxide (NaOH) method as recommended [13].
These steps may create aerosols that cause a major health risk to the operator and may also cross-contaminate specimens if appropriate containment measures are not followed. For reliable results and interpretable, we introduced in each run of DNA extraction a negative control. During the study, presence of DNA suggesting samples contamination was not observed by analysis of agarose gel stained with ethidium bromide containing the PCR products. In fact, during extraction process the biologist who performed decontamination process was different from who performed DNA extraction. A second aspect may probably be due to motivation of the human resources who were well trained. Globally, all results observed with others controls included in the LPA process were correct. The migration of PCR products is optional [13] but it is very important for implementation of Line

Table 2. Description of patients according to age group and sex.

Table 3. Susceptibility of Rifampin and Isoniazid according to categories of patients.

Table 4. Comparison of Rifampin resistance in categories of patients.
Probe Assay.
Patients of category II (treatment failures, relapse, and defaulters) are treated with the 2RHZES/1RHZE/5HRE regimen in Côte d’Ivoire as in many developing countries [14]. Among 278 sputum samples collected, 148 (53.2%) were obtained from Recurrent TB cases. The recurrence of active TB after treatment can be due to relapse (endogenous reactivation) or exogenous re-infection with a new strain. Nyamogoba et al. have reported in their study a high TB recurrence rate in Kenya independently of human immunodeficiency virus infection [15]. However, the present study could not determine whether the recurrence were due to endogenous reactivations or exogenous re-infection. Re-infection among this group is not negligible [16]. The majority of recruited patients was in the 25 - 34 year age-group, one of the most productive segments of society independently of patients category. The previously tuberculosis therapy being a risk factor for drug resistance tuberculosis, rapid identification of Mycobacteria from smear-positive sputum samples is important in term the TB control [3,4].
Many of rapid molecular methods for TB diagnosis are available [5-7]. Before their implementation for case detection, a validation is advocated. This step is important but not essential for implementation of a new diagnostic method. Concerning our study, molecular assay used, has been evaluated by different authors who showed that this method reduced time to detection of MDR-TB, and the performances (sensitivity and specificity) of the assay are acceptable for susceptibility detection to Rifampin and Isoniazid with sputum and strains [5,17,18].
Directly detection of drug resistance with sputum (positive smear) by using a molecular assay is indicated for a good interpretation of results obtained [19-21]. Considering AFB detection after Ziehl-Neelsen colouring and internal quality program for each sequence of DNA extraction, amplification and hybridization, permitted probably to reduce contamination and to get interpretable results. The Molecular method used has permitted to detect simultaneous resistance to Rifampin and Isoniazid from sputum of 148 (53.2%) previously treated patients.
Considering each patient category, it appears that multidrug-resistant tuberculosis (Resistance Rifampin and Isoniazid) changes according to patient category. Indeed, in cases of failure, rates observed were 69.8% (95% IC, 60% - 80%), 75% (95% IC, 60% - 90%) respectively for 2RHZE/4RH regimen and 2RHZES/1RHZE/5HRE regimen (Table 3). In several countries (Russia, South Africa, Philippines) high rates of Rifampin and Isoniazid resistance have been described in adults [3]. The high rate of resistance observed in previously treated patients with 2RHZE/4RH regimen could be one reason of ineffectiveness of retreatment described by [22] in Côte d’Ivoire.
Rifampin resistance is a good, but not, a perfect marker for multidrug-resistant tuberculosis [23]. Used as a marker of MDR-TB, study showed that there was no statistically significant difference for MDR-TB cases observed in failures cases (Table 4). The observation suggests that to establish an appropriated treatment regimen the drug susceptibility testing is required to reduce resistant strains propagation in the community. Concerning recurrent TB cases, 69 were resistant at least to Rifampin (Table 4). Compared to those observed in failures cases, difference is statistically significant. Indeed, in case of treatment failures, one of main reason is probably an infection with multidrug-resistant strains. Whereas in recurrent TB case, infectious strains are susceptible or resistant.
5. Conclusion
In Côte d’Ivoire, genotype® MTBDRplus assay has permitted to estimate the prevalence of MDR-TB in categories of previously treated patients for tuberculosis.
6. Acknowledgements
We thank Natalia Shubladze the Consultant of American Society for Microbiology Dr. Daniela Cirillo of SRL of Milan and KEKELETSO Kao of Expand TB for their contributions. We also thank the Médecins-Chefs of Centre Antituberculeux in Côte d’Ivoire.