Cytogenetic and Molecular Characterization of Hematological Neoplasm in an Ecuadorian Population ()
1. Introduction
Hematological disorders are an important cause of morbidity and mortality in the entire world. A database produced by the Leukemia and Lymphoma Society—USA (updated in 2013), shows the following rates per 100,000 inhabitants worldwide: 12.8 for leukemia, 19.7 for NonHodgkin lymphomas (NHL), 2.8 for Hodgkin’s lymphoma (HL), and 5.9 for myeloma [1].
The equivalent in Ecuador is produced by the National Cancer Registry of SOLCA [2], located in the city of Quito (capital of Ecuador), which reports that in the period of 2001 to 2005 tumors of the hematopoietic and reticuloendothelial systems are within the 25 most frequent malignancies, occupying the fourth place (5.5%) in males, and the sixth place (4.5%) in females. During the 2003-2005 period, analysis of the average annual incidence of their malignancies shows a crude rate of 9.3 (5.3%) and 9.1 (4.6%) in males and females respectively. Hodgkin’s lymphoma (HL) has a crude rate of 1.4 and 0.4, follicular (nodular) NHL 0.8 and 0.9, diffuse NHL 4.7 and 3.9, peripheral and cutaneous t-cell lymphomas 0.8 and 0.7 and other types of NHL 3.2 and 3.0 in males and in females, respectively. The crude rate for Multiple Myeloma (MM) and plasma cell neoplasms, in the same period of time is 2.1 for males and 1.5 for females, for lymphoid leukemia (LL) is 3.9 and 4.3, for myeloid leukemia (ML) is 2.5 and 2.7, for monocytic leukemia is 0.1 in males and for leukemia of unspecified cell type is 0.6 and 0.5, in males and females respectively, finally the myelodysplastic syndrome (MDS) shows a crude rate of 0.1. Hematopoietic and reticuloendothelial system malignancies have an average annual mortality rate of 5.0 in males and 4.4 in females per 100,000. According to the relative position of the 25 most frequent malignant tumors in males, in the period 2001-2005, lymphoma (6%) and leukemia (4%) are in the fourth and fifth places respectively, with MM and malignant plasma cell neoplasms in the 21st place with 1.1%. Data in females differs slightly, showing 4.3% in lymphoma (sixth place), 3.6% leukemia (eighth place) and MM and malignant plasma cell neoplasms in the 25th place with 0.6%. LL (56%) has been reported as the most common in the general population of Quito and ML is 36%. The latter, is more frequent in adults and represents 58%. Regarding lymphomas, the most frequent is diffuse NHL (44%), followed by follicular NHL (10%), HL (10%) and finally T cell Lymphoma, peripheral and cutaneous lymphomas (6%) [2]. A study among indigenous people in the Amazon region of Ecuador, reported that leukemia was one of the most common type of cancer in that population [3].
Ecuador’s data are also reported by the International Agency for Research in Cancer, through the GLOBOCAN project, showing an estimated incidence (2008) of crude rates for leukemia at 6.1, 5.1 for NHL, 0.9 for MM and 1.2 for HL; therefore it would be interesting to compare these results to worldwide data which reports crude rates of 5.2 in leukemia, 5.3 in NHL, 1.5 in MM and 1.0 in HL [4].
Acute myeloid leukemia (AML) is the most common leukemia in adults. The main genetic abnormalities related to this disorder are: t(8;21), inv(16) and t(16;16) and t(15;17) [5], which have been reported in other studies with incidences of 0 to 4% [6,7]. Rearrangements of chromosome 11 (11q23) are observed in 4% - 10% of AML patients [8]. The following three new rearrangements related to AML have also been reported: t(6;9), inv(3) or t(3;3) and t(1;22). Regarding chromosomal numerical aberrations, trisomy 8 is the most common in AML [5].
Acute lymphoblastic leukemia (ALL) is the most frequent hematological disorder in children, with t(12;21) being the most frequent abnormality [9]. The second most frequent alteration is t(9;22), otherwise known as Philadelphia chromosome (Ph) which present in approximately 5% of children and 20% of adults with ALL [10].
So far few studies have analyzed the incidence of molecular alterations in ALL within Latin America. Frequency analysis for the MLL-AF4, TEL-AML1 and BCR-ABL genes in children from Mexico City, reported 65.4% for the first rearrangement and 3.8% for the two remaining [11]. In a Peruvian population PML-RARα presence was analyzed in patients with acute promyelocytic leukemia (APL) and the results showed that the bcr1 subtype was present in 62% of individuals [12]. Another study performed on a group of patients also originated from Latin America showed 75% for bcr1, 10% for bcr2 and 15% for bcr3 [13].
Chromosome Ph is considered the initial event in Chronic Myelogenous Leukemia (CML), as the result of the BCR-ABL gene fusion [14]; it was reported that 92% of CML patients were positive for the BCR-ABL gene, specifically b3a2 (61%) and b2a2 (31%) [15]. However, frequencies of the different isoforms in Ecuador vary and are as follows: b2-a2 (94.6%) y b3-a2 (5.4%) in CML [16]. Atypical b3a3 rearrangement was also described by our group in a patient with a good response to INFalpha treatment [17].
There is not a registry of genetic alterations of the hematological disorders in Ecuador. Therefore, this study is the first to combine information from three reference centers, which perform genetic analyses in samples of patients from all over the country.
The main objective of this study is to delineate a genetic database of hematological disorders that affect the Ecuadorian population and at the same time compare our results to studies from other countries. This group of blood disorders is characterized by geographic heterogeneity of chromosomal and molecular abnormalities. These regional variations, as well as variation in the types of medical practices, quality of treatment and age (which may differ amongst different cities) influence the frequency of cancer diagnoses and patients’ deaths. [18- 20]. The combination of data will help achieve more accurate diagnostics, through the interchanges of cytogenetic and molecular findings that complement each other, which will improve the treatment and management of patients with leukemia and lymphoma.
2. Methods
We analyzed 4108 patients with various hematological malignancies, between 1984 and 2012, who were referred to the genetic study from ten institutions from different cities of Ecuador. The hematological diseases were diagnosed according to the WHO classification.
Cytogenetic analysis was performed in bone marrow cells immediately after collection. G-banding technique was performed using standard approaches, and was aimed at identifying structural and numerical alterations, which were classified according to the International System for Human Cytogenetic Nomenclature [21].
Cytogenetic results were classified into five subgroups: Normal karyotype, hyperdiploid, hypodiploid, translocation and complex karyotype. The translocation group included t(8;21), t(15;17), 11q23 rearrangements, t(1;19) and t(9;22) that are specifically associated with the ALL, AML and CML. The presence of three or more chromosomal alterations and the non-frequent chromosomal abnormalities in the hematopoietic malignancies of this study were classified as complex karyotype group.
Patients were divided into two age groups. The pediatric group which consisted of patients ≤15 years, and the adult group of patients >16 years of age [22,23].
The fusion genes were detected using the t(8;21) (q22;q22), t(15;17) (q21;q22), inv(16)/t(16;16)(p13;q22), t(9;22)(q34;q11.2), t(1;19)(q23;p13.3), t(12;21) (p13;q22) and translocations involving 11q23 and 8q24.1 probes (Vysis RUNX1/RUNX1T1 DF FISH Probe Kit; Vysis LSI PML/RARA Dual Color Single Fusion Probes; Vysis LSI CBFB Break Apart Rearrangement Probe; Vysis LSI BCR/ABL Dual Color, Single Fusion Translocation Probe; LSI TCF3/PBX1 Dual Color, Dual Fusion Translocation Probe; Vysis LSI ETV6(TEL)/ RUNX1(AML1) ES Dual Color Translocation Probe Set; Vysis LSI MLL Dual Color, Break Apart Rearrangement Probe; and Vysis LSI MYC Dual Color Break Apart Rearrangement Probe, respectively) according to the manufacturer’s instructions (Abbott). Two hundred cells were analyzed for each sample, and normal blood cells were used as a negative control.
Total and messenger RNA was extracted from bone marrow samples, using methods previously described [24]. Analysis by reverse transcriptase-polymerase chain reaction of the RNA of the samples was carried using nested primers defining the junction site of the chimeric gene AML1-ETO, PML-RARA, CBFB-MYH11, BCR-ABL, E2A/PBX1 and AF4-MLL using primers previously described [25].
3. Results
3.1. Clinical Data
Out of the 4108 newly diagnosed patients with different hematological malignancies, 2326 (56.7%) patients were male and 1780 (43.3%) patients were female. Patient age ranged from one month to 87 years, with a median age of 43 years. The different malignancies included 715 AML, 1260 ALL, 168 MDS, 52 myeloproliferative disorder (MPD), 945 CML, 15 polycythemia vera (PV), 63 essential thrombocythemia (ET), 104 chronic lymphocytic leukemia (CLL), 10 hairy cell leukemia (HCL), 4 monoclonal gammopathy of undetermined significance (MGUS), 165 MM, 2 plasma cell leukemia (PCL), 1 Waldenstrom macroglobulinemia (WM), 14 medular aplasia, 5 Fanconi anemia (FA), 80 HL, and 505 NHL cases (Figure 1). These cases were categorized by age and sex and are shown in Figures 2 and 3.
3.2. Conventional Cytogenetics
We used conventional cytogenetics to analyze a minimum of 20 metaphases per patient in 4108 patients. As

Figure 1. Types of hematological diseases in which genetic study was conducted in Ecuadorian population, 1984-2012.

Figure 2. Hematological diseases analyzed by group of age in the Ecuadorian population.

Figure 3. Hematological diseases analyzed by gender in the Ecuadorian population.
shown in Table 1, chromosome abnormalities were detected in 1886 (45.9%) patients, whereas 1405 (34.2%) patients had a normal karyotype. Chromosome abnormalities included numeric chromosome changes (24.6% patients), structural chromosome changes (50.6% patients), and complex karyotypes (24.8% patients). The rest of the samples were not in metaphase and therefore were not classified by this technique. Within the struc-

Table 1. Cytogenetics findings in Ecuadorian patients with hematological neoplasm.
tural chromosomal changes the most common translocations were t(8;21), t(15;17), 11q23 rearrangements, t(1; 19) and t(9;22) and were associated hematologic neoplasms.
The most frequent chromosomal alterations reported in leukemia were also detected in this study. Some of the most important found are the following:
In AML the main chromosomal alterations found were t(8;21) (4.3%), t(15;17) (3%), inv(16) (1.3%), t(9;11) (1%), trisomy 8 (4%) and other translocations (5.3%). The most frequent in ALL were t(9;22) (15.5%), t(1;19) (4.8%), t(4;11) (1.2%), 11q23 rearrangements and t(8;14) (0.4% each). t(9;22) was detected in 80% of CML cases. In MM cases, 8.5% of the cases showed rearrangements of chromosome 14. In NHL 20% of the altered metaphases showed translocations.
3.3. FISH Evaluation
FISH was performed in samples of patients without metaphases or with normal karyotype; the probes used were those of the most frequent rearrangements in leukemia. We found alterations in 13/715 AML patients, 31/1260 ALL patients and 7/945 CML patients, which increased our positive results by 2%, 2.5% and 1% respectively (Table 2).
3.4. Molecular Study
The results obtained by FISH were consistent with the results of molecular analysis (Table 2). In cases without metaphases, alterations were detected in 22/715 AML patients, 29/1260 ALL patients and 65/945 CML patients, which increased positive results in 3.1%, 2.3% and 6.9% respectively comparing to conventional cytogenetics results.
Cases in which t(8;21) was detected by FISH also showed the AML1-ETO RT-PCR product. AML patients with PML-RARA rearrangements presented the bcr2 transcript (36%) as well as the bcr3 transcript (64%), however, none of them presented the bcr1 transcript. The CBFB-MYH11 fusion transcript was present in 7 patients, all diagnosed with ALL; all of these the presented the type F transcript.
The BCR-ABL fusion gene was detected using RTPCR in all t(9;22) positive ALL cases; all of the aforementioned samples showed the e1-a2 transcript. Among the 11q23 rearrangements found in 10 patients
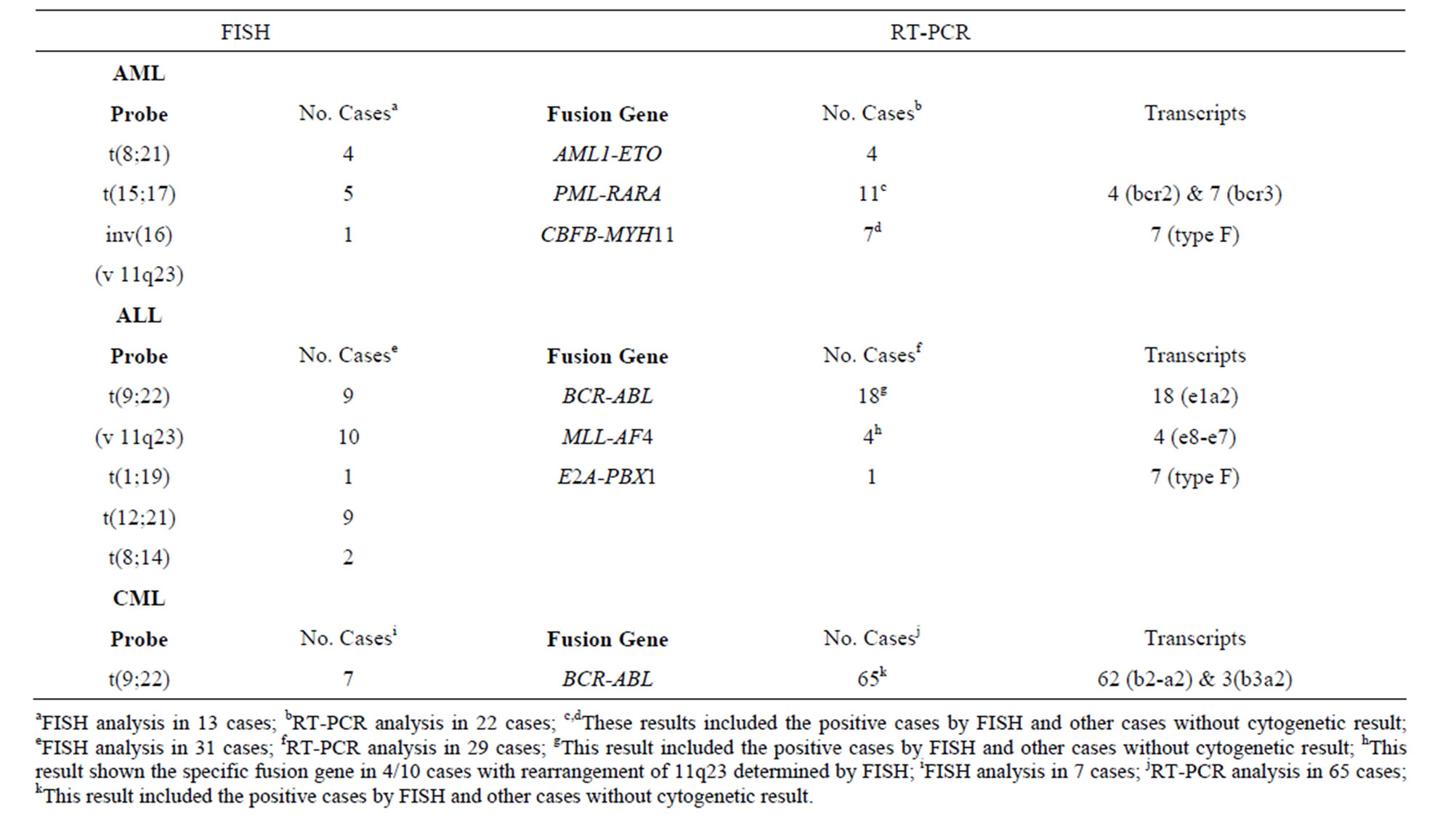
Table 2. Positive results in AML, ALL and CML cases without metaphases and normal karyotypes.
with ALL, 4 corresponded to the t(4;11) within the MLL-AF4 fusion gene, in which the transcript e7-e8 was observed in all cases. The t(1;19) presented the E2APBX1 fusion gene. We identified b2a2 (95%) and b3a2 (5%) fusion transcripts in CML patients carrying the BCR-ABL fusion gene.
In AML patients with the 11q23 rearrangements and ALL patients with t(12;21) and t(8;14) we did not perform RT-PCR analysis.
4. Discussion
The incidence of hematological disorders in Ecuador constitutes a major cause of mortality. Geographical heterogeneity was described regarding chromosomal alterations and molecular hematologic problems but the information in Ecuador is limited.
Our group has previously reported on the frequency of BCR-ABL rearrangements in CML and ALL [16,17] and the incidence of follicular lymphoma and t(14;18) [26] in Ecuador. Within this study we present the chromosomal characterization of patients with hematologic disorders who were referred to the genetic analysis, which provides a better understanding of the evolution and prognosis of afflicted individuals. We present data from genetic studies conducted in three reference centers of Quito since 1984, when cytogenetic studies began in Ecuador. These centers analyzed their patients as well as patients referred from ten institutions of various Ecuadorian cities.
Numerical and structural chromosomal abnormalities were detected in 45.9% of individuals; this is significant in the prognosis of each patient and to their specific response to certain therapeutic agents. In cases where we could not obtain a karyotype, or a normal karyotype was obtained, FISH probes were applied. This allowed us to reveal an average of 1.8% more alterations, therefore increasing the number of patients classified within each type of blood disease. These alterations were mainly specific translocations associated to leukemia. FISH positive cases were validated by analyzing fusion genes using RT-PCR. All cases positive for AML, ALL and CML presented the following translocations: t(8;21), t(15;17), inv(16), t(9;22) and t(1;19). These translocations exhibited the following RT-PCR products respectively: AML1- ETO, PML-RARA, CBFB-MYH11, BCR-ABL and E2APBX1. Studies have identified at least 20 partners with translocations involving the MLL gene (11q23) [27]. Within this study only forty per cent of the rearrangements of 11q23 as evidenced by FISH corresponded to t(4;11) with the MLL-AF4 fusion gene.
Using RT-PCR allowed us to increase the genetic alterations we observed in the samples analyzed by approximately 4.1%. In the case of CML patients, the Ph chromosome was detected in 80% of individuals analyzed by conventional cytogenetics. FISH identified the t(9;22) in cases without metaphases, bringing the cases with CML up to 81%. BCR-ABL detection using RTPCR analysis helped increase the detection of Ph chromosome in 86.9% of CML cases, representing a significant increase compared to the cytogenetic results.
While this study has focused in the molecular and cytogenetic characterization of hematological neoplasms in an Ecuadorian population, we also examined the frequencies of the different transcript types of fusion genes. Variations in breakpoint sites were correlated to the phenotype and malignancy of leukemia [28]. Cases with t(8; 21) (as detected by FISH) showed exclusively the presence of the AML1-ETO fusion gene using RT-PCR. In the case of t(1;19), again identified by FISH, the E2APBX1, showed the most common RT-PCR product.
Rearrangement frequencies of the subtypes b2-a2 (95%) and b3a2 (5%) of BCR-ABL in CML were statistically different to the frequencies reported in other populations [29] and are similar to previously reported data from Ecuadorian patients [16]. In contrast, the e1-a2 rearrangement of the BCR-ABL gene, present in approximately 95% of ALL patients [25], was detected in 100% of positive cases in this study. The e8-e7 transcript was visualized in all cases positive for the MLL-AF4 fusion gene. This variant is rare in non-infants and has been reported in less than 10% of infants. Positive cases had a median age of 7 years (range, 2 - 12 years). The detection of this fusion gene in ALL infants is particularly important because they have a poor response to chemotherapy [30].
The transcripts of the PML-RARA fusion gene corresponded to bcr2 (36%) and bcr3 (64%). These results are markedly different from the data reported by other groups: bcr1 (55%), bcr2 (5%) and bcr3 (40%) [31]. In our population, none of the AML patients presented the bcr1 variant. The presence of a particular variant in a patient or a population can be of particular importance as it could have an effect on treatment response and final outcome [32].
CBFB-MYH11 is associated with a good prognosis [33]. All ALL patients with this fusion gene showed the type F transcript, however in general few cases have been described with this kind of gene transcript. The most frequent transcript is type A, present in more than 85% of positive cases. Transcripts D and E, represent about 5%, while others are unique cases [34]. It is noteworthy that although the number of positive cases by RT-PCR was reduced, all were of type F.
The differences in the frequencies of some fusion genes may be a response to the type of the population analyzed. Ecuador has 500 years of history of a mixture between mainly Native Amerindians and Spanish populations. It must also be considered that the differences in the frequency of the different transcripts can be the result of a different genetic component in the Ecuadorian population compared to other populations and that has been observed in studies, reported by our group, involving repair genes associated with NHL and CML [35,36]. FISH analysis and RT-PCR methods were used in cases which could not be resolved by conventional cytogenetics in order to obtain results that contribute to patients’ diagnosis. The differences in the frequencies of the transcripts of some fusion genes show a different genetic behavior similar to previously described genes related to diseases such as ΔF508 in cystic fibrosis, hRAD54 in meningioma and HFE in hemochromatosis [37-39]. Thus, in-depth studies of fusion genes in different hematological malignancies permit a better understanding of the presence of different transcripts and their effect on leukemia.
5. Acknowledgements
We would like to thank Melissa Arévalo (HCAM), Catherine Carrera (SOLCA), Mirian Espín (SOLCA), Alicia Godoy (HCAM) and Janeth Nájera (HCAM) for technical assistance.